Detailed DSSR results for the G-quadruplex: PDB entry 1jb7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1jb7
- Class
- DNA-binding protein-DNA
- Method
- X-ray (1.86 Å)
- Summary
- DNA g-quartets in a 1.86 Å resolution structure of an oxytricha nova telomeric protein-DNA complex
- Reference
- Horvath MP, Schultz SC (2001): "DNA G-quartets in a 1.86 A resolution structure of an Oxytricha nova telomeric protein-DNA complex." J.Mol.Biol., 310, 367-377. doi: 10.1006/jmbi.2001.4766.
- Abstract
- The Oxytricha nova telomere end binding protein (OnTEBP) recognizes, binds and protects the single-stranded 3'-terminal DNA extension found at the ends of macronuclear chromosomes. The structure of this complex shows that the single strand GGGGTTTTGGGG DNA binds in a deep cleft between the two protein subunits of OnTEBP, adopting a non-helical and irregular conformation. In extending the resolution limit of this structure to 1.86 A, we were surprised to find a G-quartet linked dimer of the GGGGTTTTGGGG DNA also packing within the crystal lattice and interacting with the telomere end binding protein. The G-quartet DNA exhibits the same structure and topology as previously observed in solution by NMR with diagonally crossing d(TTTT) loops at either end of the four-stranded helix. Additionally, the crystal structure reveals clearly visible Na(+), and specific patterns of bound water molecules in the four non-equivalent grooves. Although the G-quartet:protein contact surfaces are modest and might simply represent crystal packing interactions, it is interesting to speculate that the two types of telomeric DNA-protein interactions observed here might both be important in telomere biology.
- G4 notes
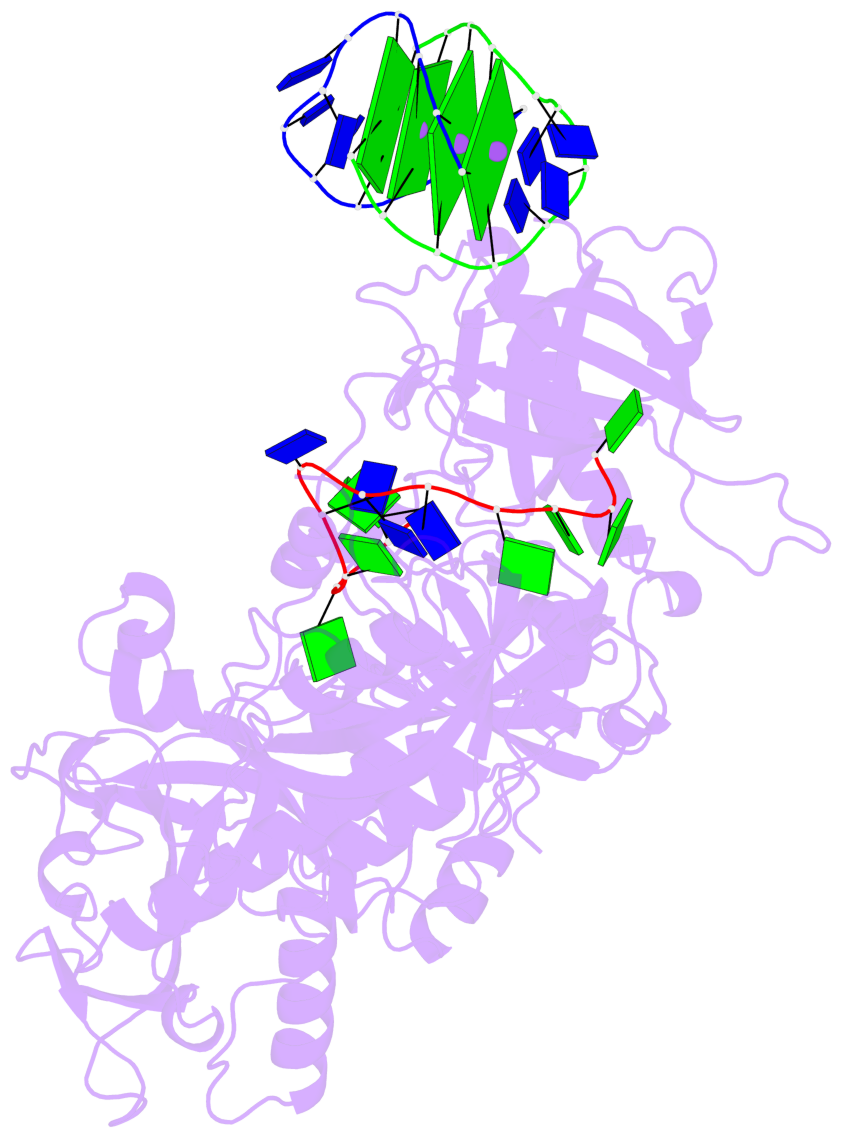
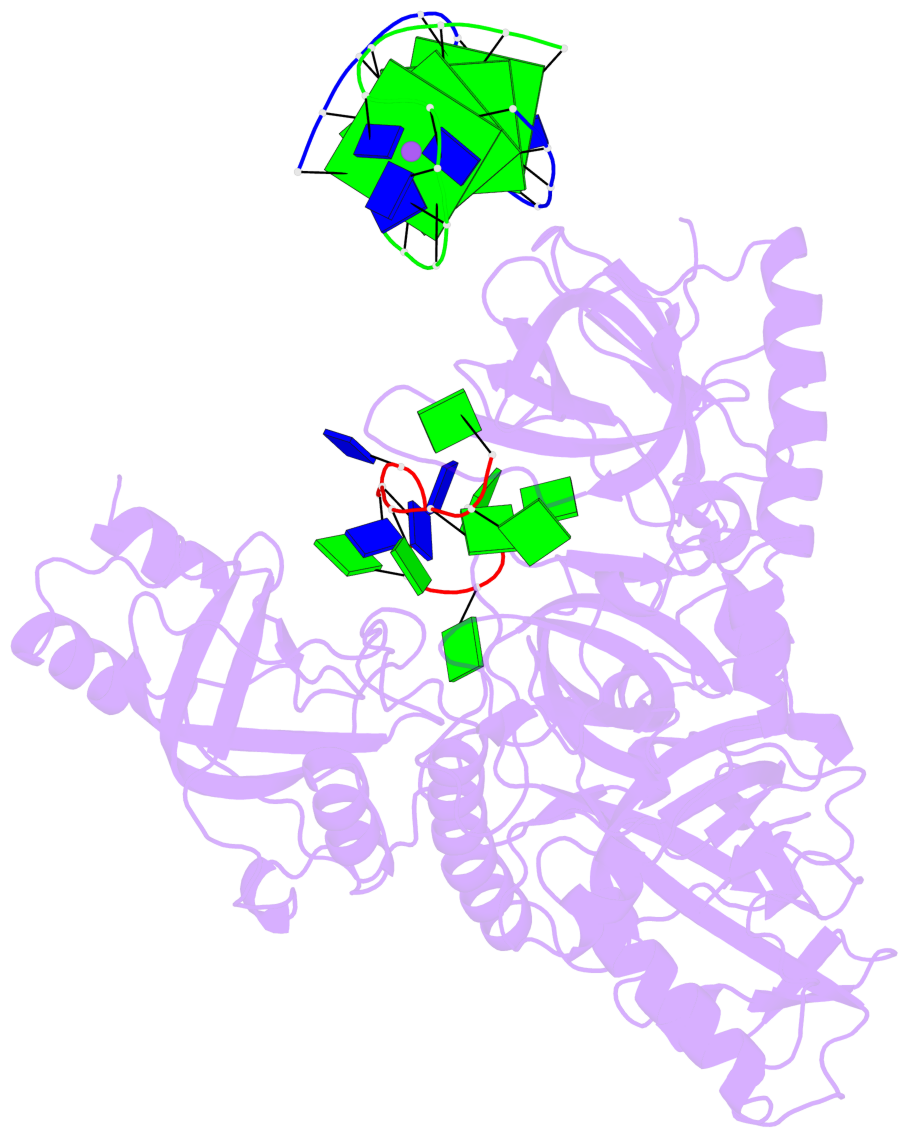
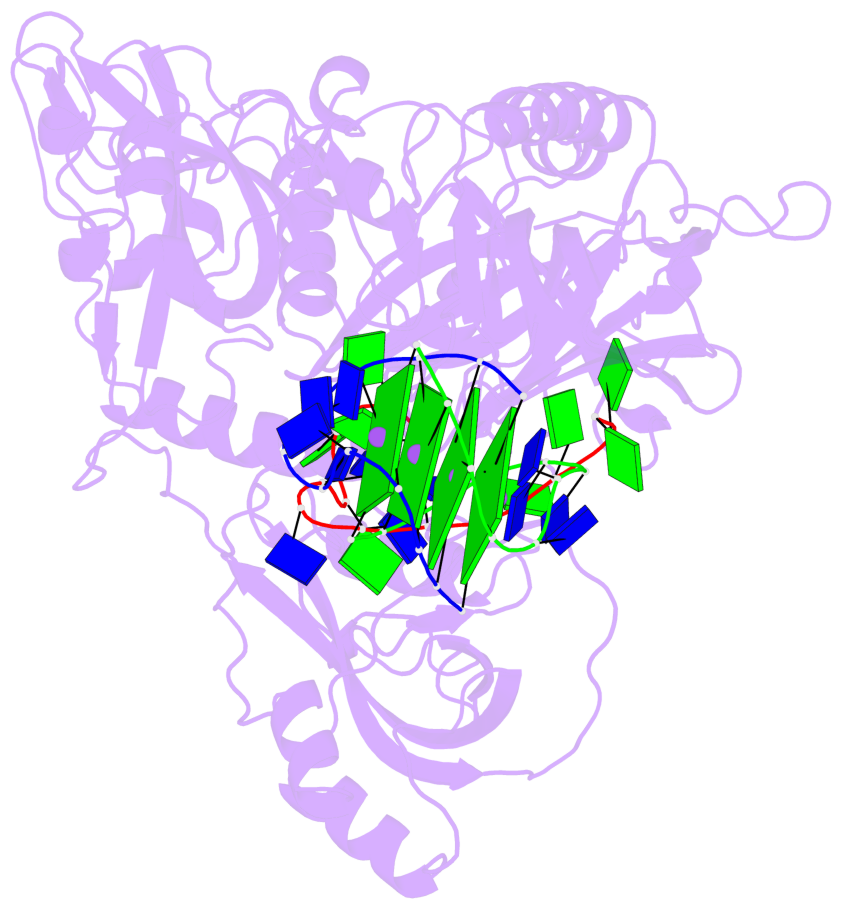
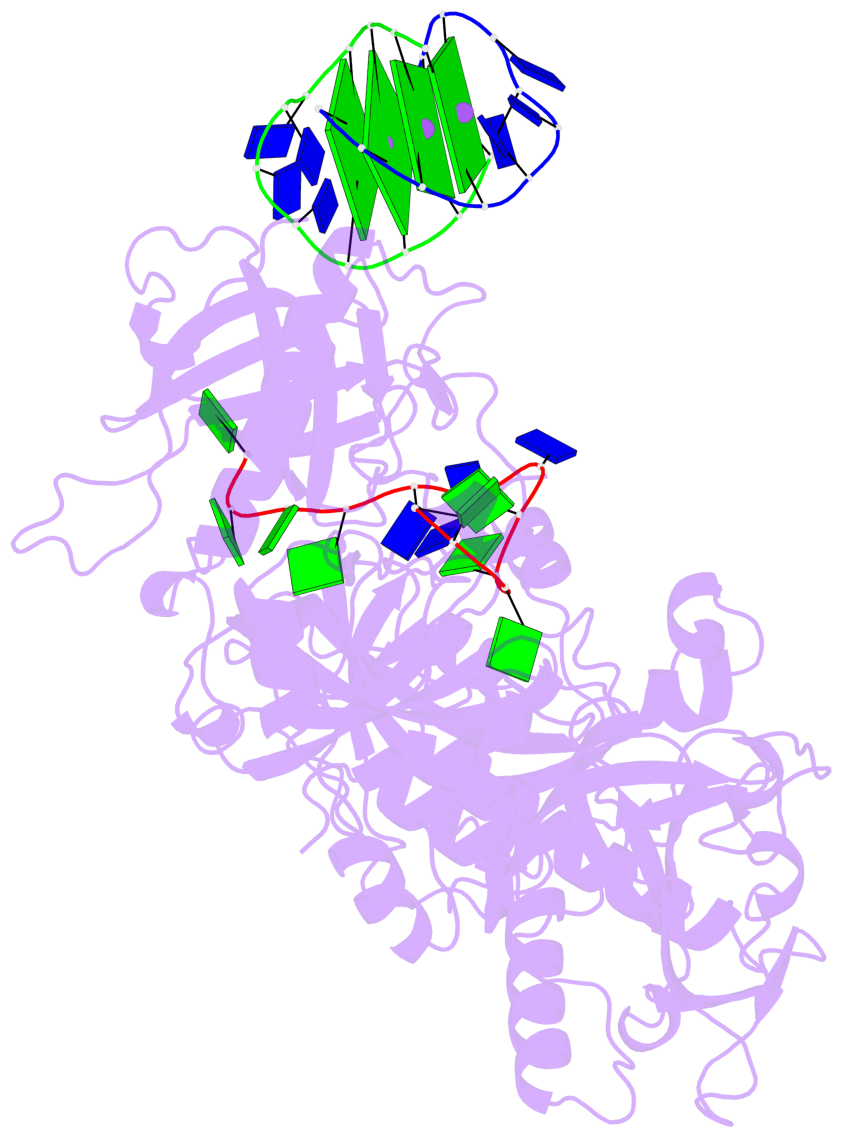
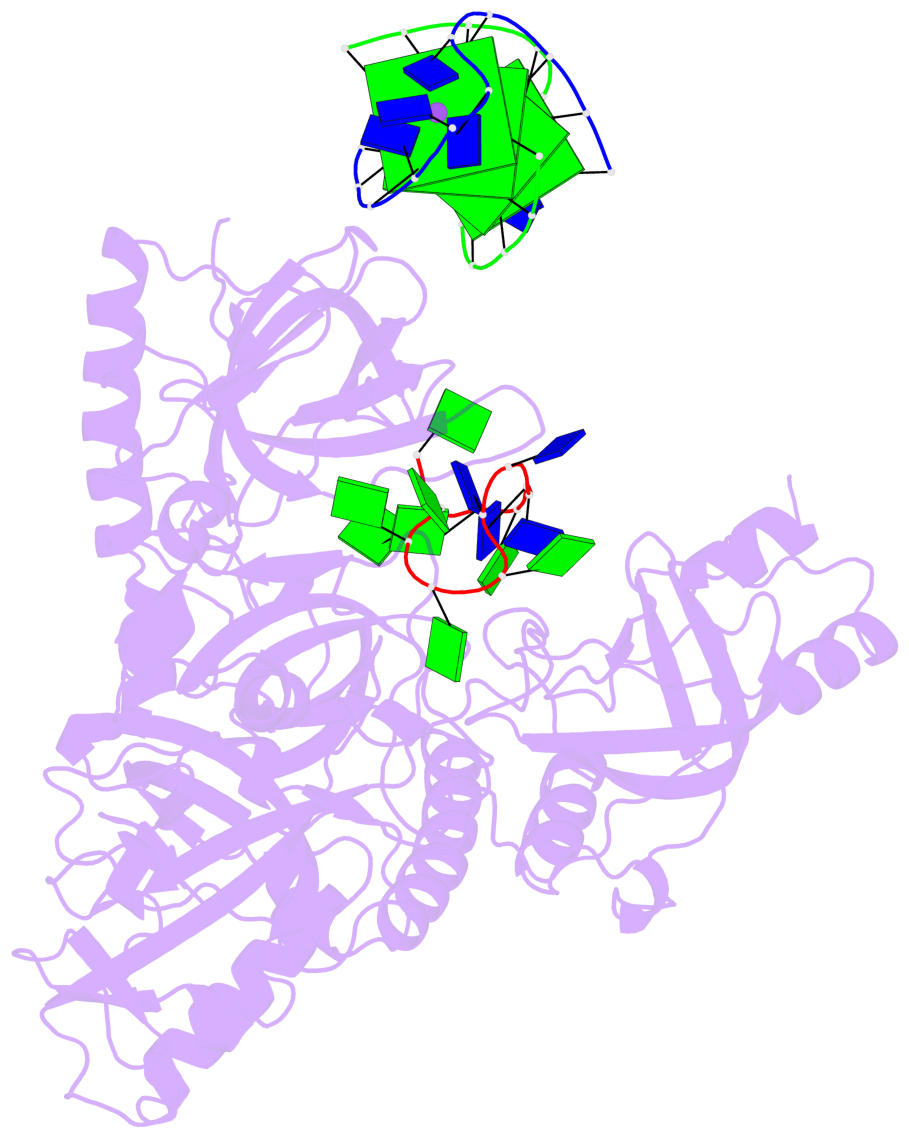
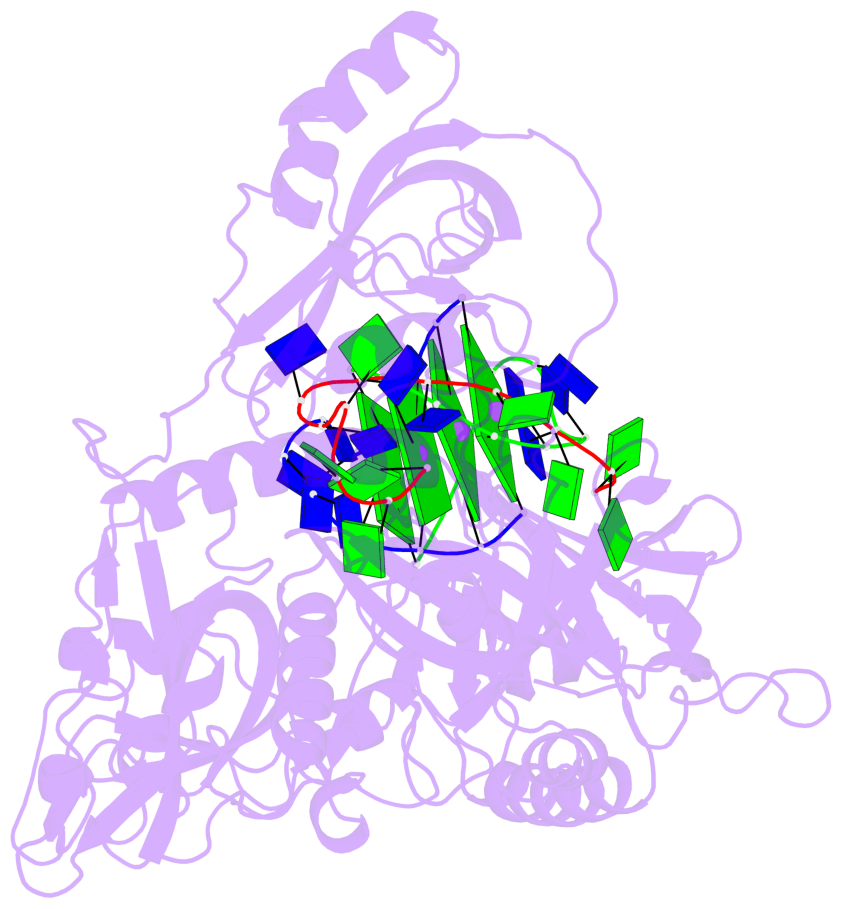
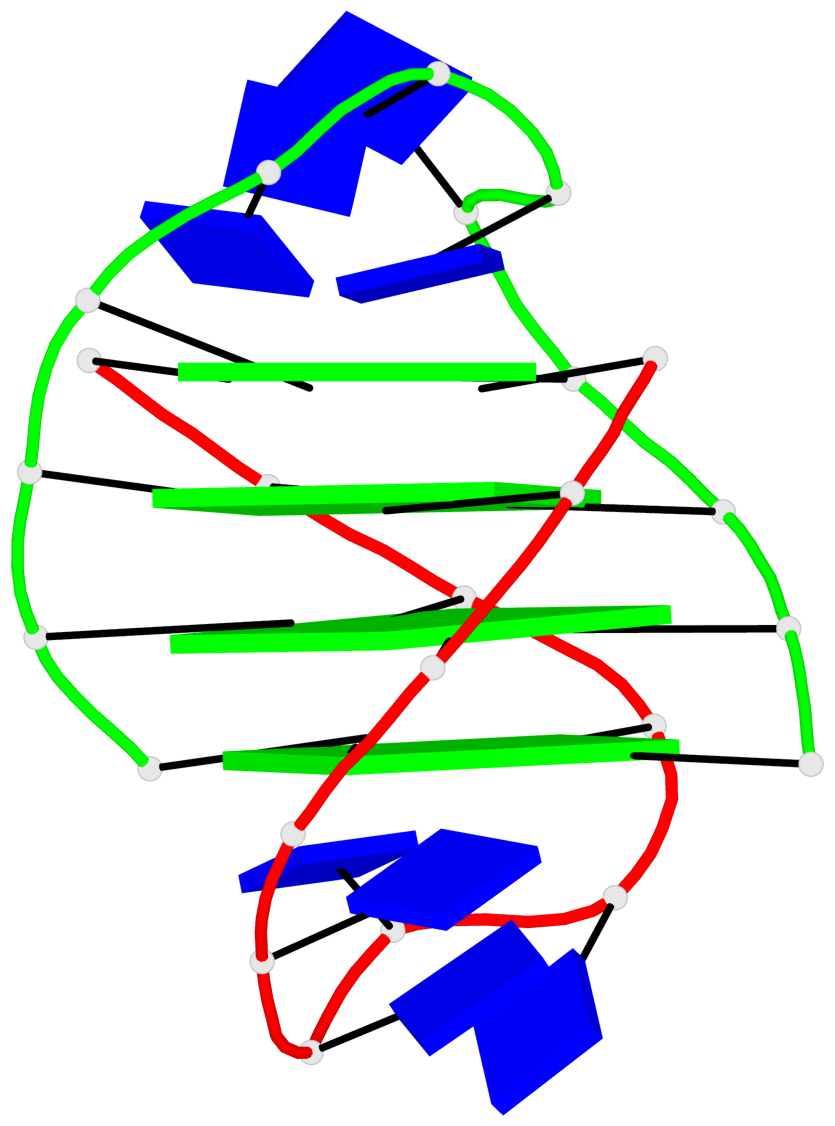
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
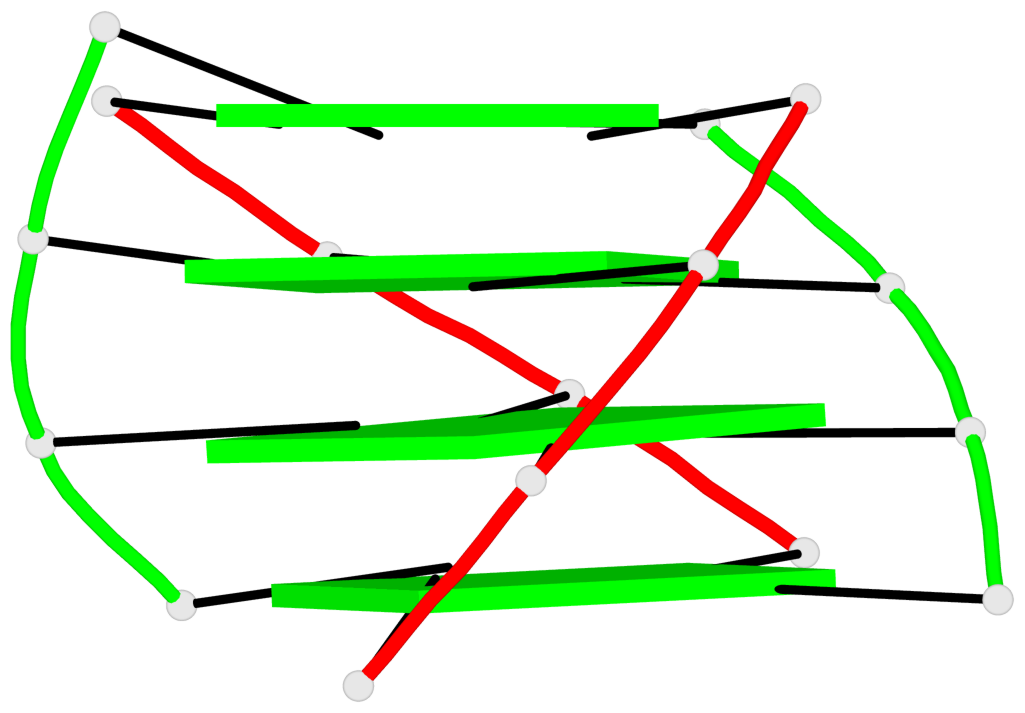
Base-block schematics in six views
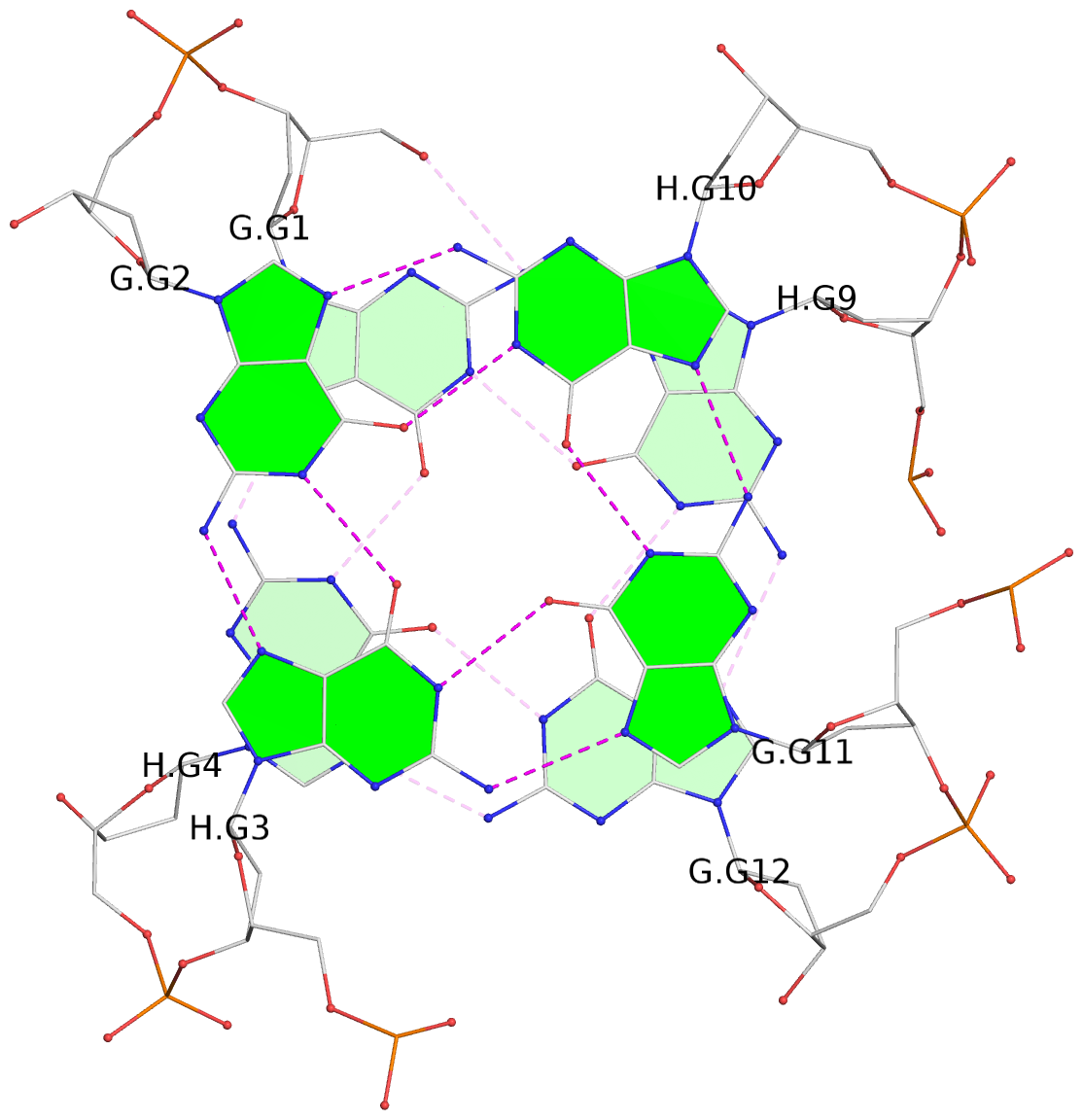
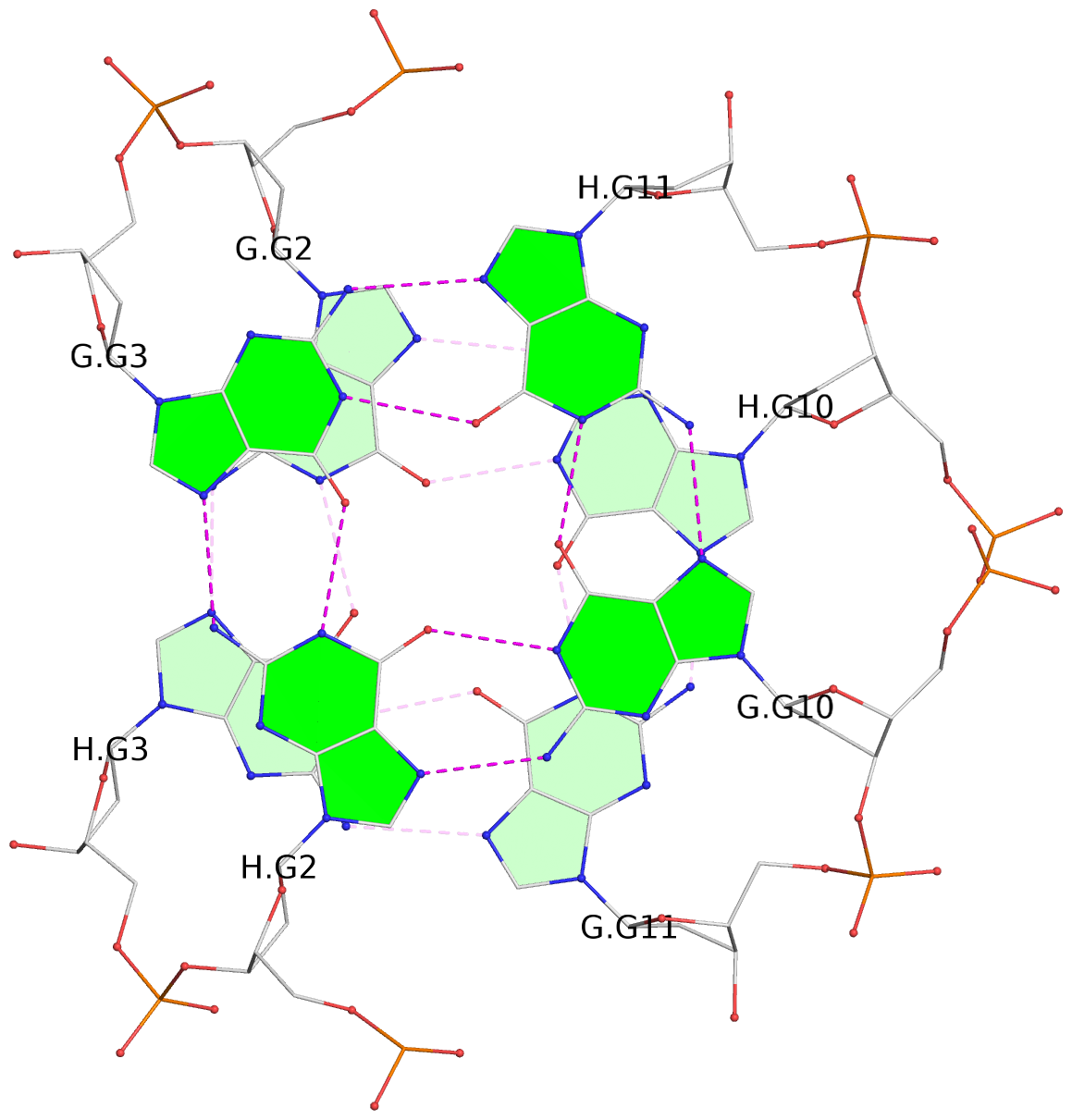
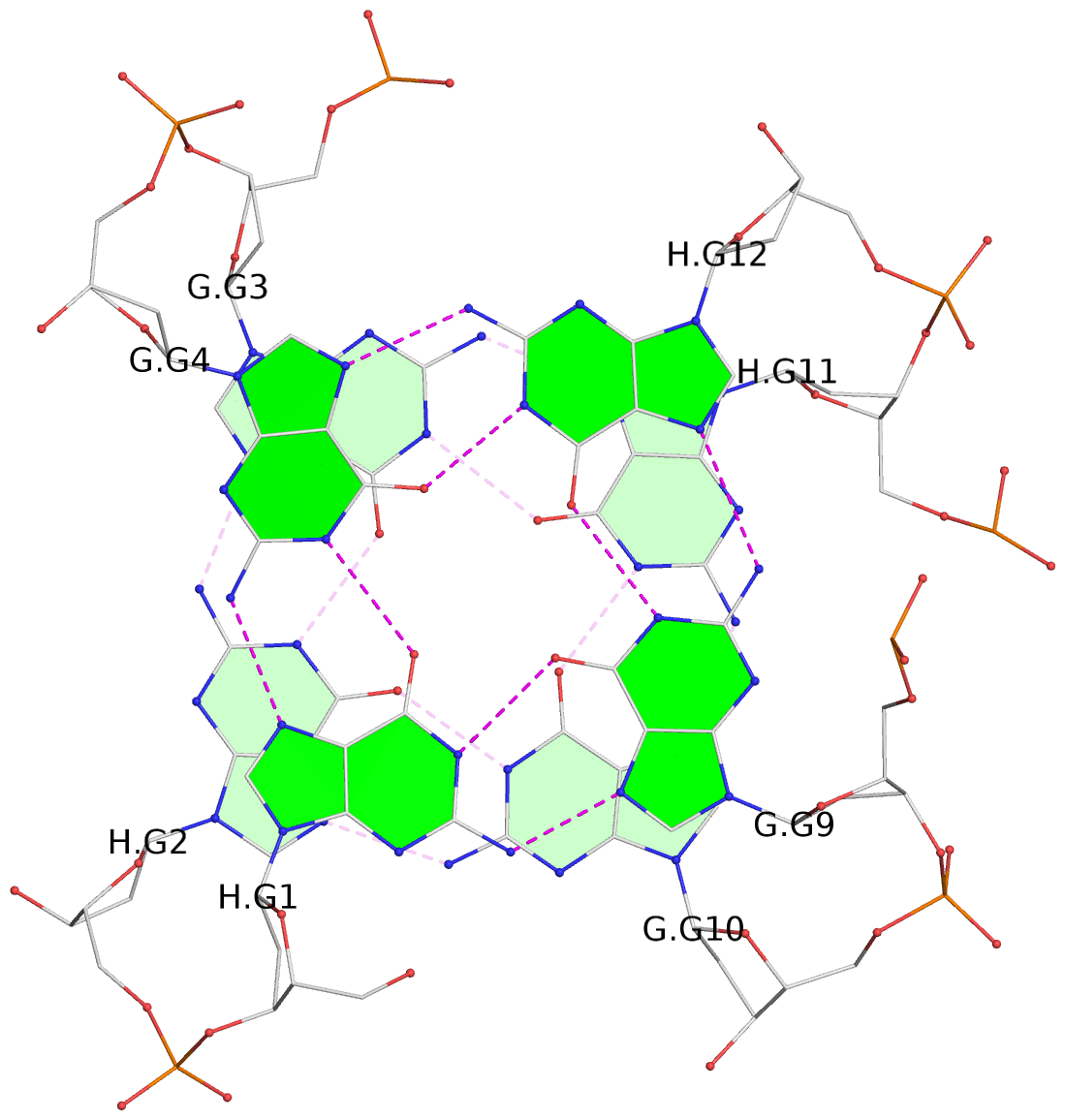
List of 4 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.321 type=bowl nts=4 GGGG G.DG1,H.DG4,G.DG12,H.DG9 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.168 type=other nts=4 GGGG G.DG2,H.DG3,G.DG11,H.DG10 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.157 type=planar nts=4 GGGG G.DG3,H.DG2,G.DG10,H.DG11 4 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.287 type=bowl nts=4 GGGG G.DG4,H.DG1,G.DG9,H.DG12
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.