Detailed DSSR results for the G-quadruplex: PDB entry 2f8u
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2f8u
- Class
- DNA
- Method
- NMR
- Summary
- G-quadruplex structure formed in human bcl-2 promoter, hybrid form
- Reference
- Dai J, Chen D, Jones RA, Hurley LH, Yang D (2006): "NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region." Nucleic Acids Res., 34, 5133-5144. doi: 10.1093/nar/gkl610.
- Abstract
- BCL2 protein functions as an inhibitor of cell apoptosis and has been found to be aberrantly expressed in a wide range of human diseases. A highly GC-rich region upstream of the P1 promoter plays an important role in the transcriptional regulation of BCL2. Here we report the NMR solution structure of the major intramolecular G-quadruplex formed on the G-rich strand of this region in K+ solution. This well-defined mixed parallel/antiparallel-stranded G-quadruplex structure contains three G-tetrads of mixed G-arrangements, which are connected with two lateral loops and one side loop, and four grooves of different widths. The three loops interact with the core G-tetrads in a specific way that defines and stabilizes the overall G-quadruplex structure. The loop conformations are in accord with the experimental mutation and footprinting data. The first 3-nt loop adopts a lateral loop conformation and appears to determine the overall folding of the BCL2 G-quadruplex. The third 1-nt double-chain-reversal loop defines another example of a stable parallel-stranded structural motif using the G3NG3 sequence. Significantly, the distinct major BCL2 promoter G-quadruplex structure suggests that it can be specifically involved in gene modulation and can be an attractive target for pathway-specific drug design.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
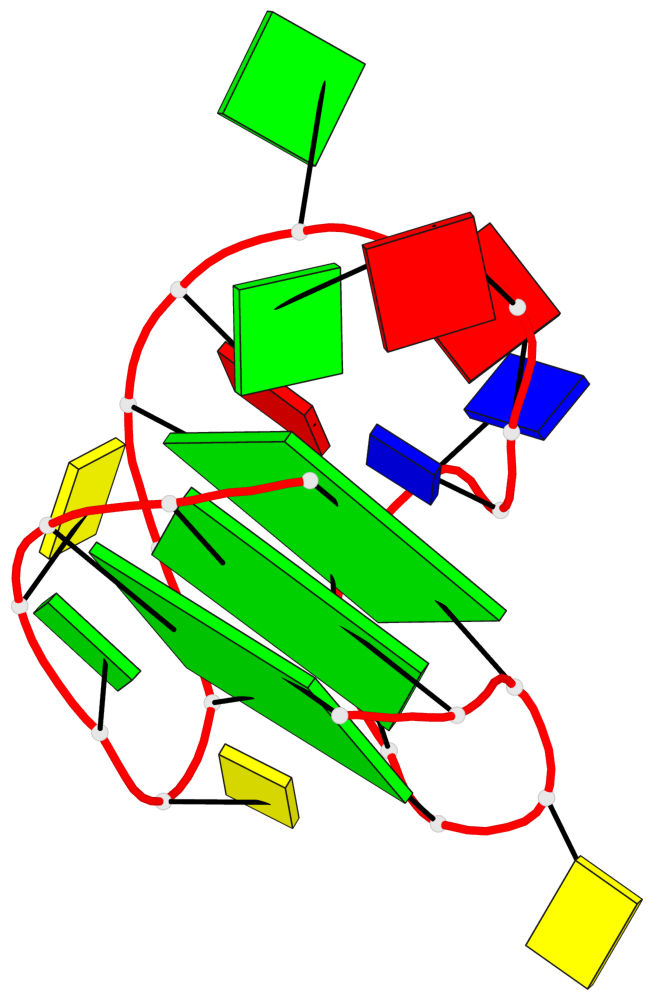
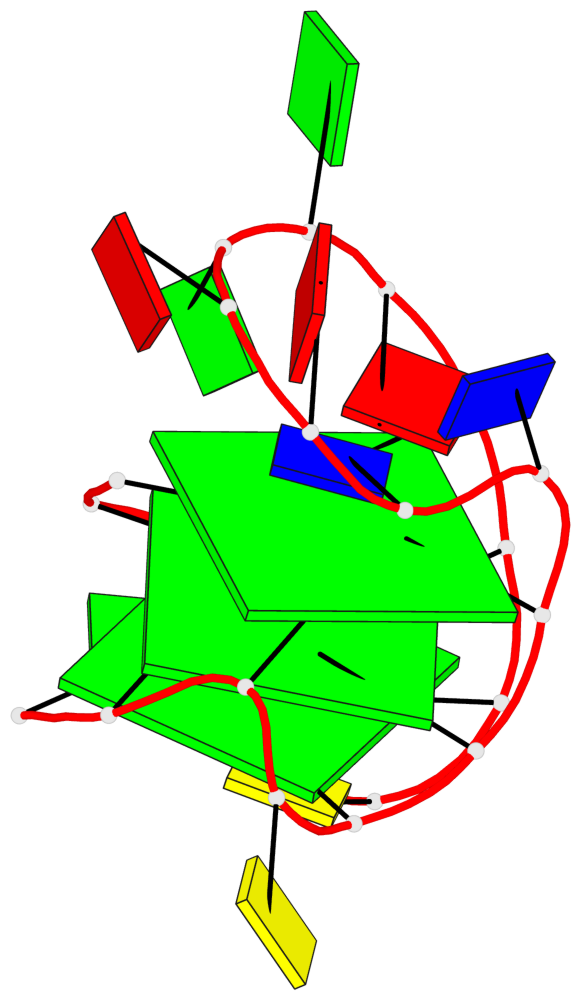
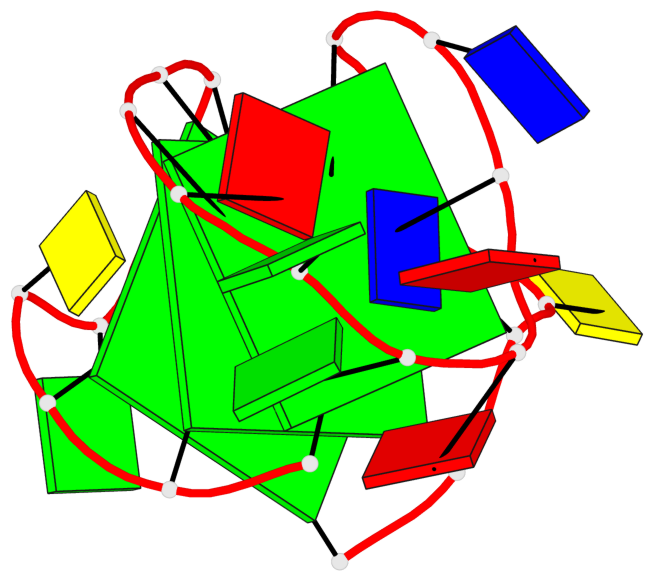
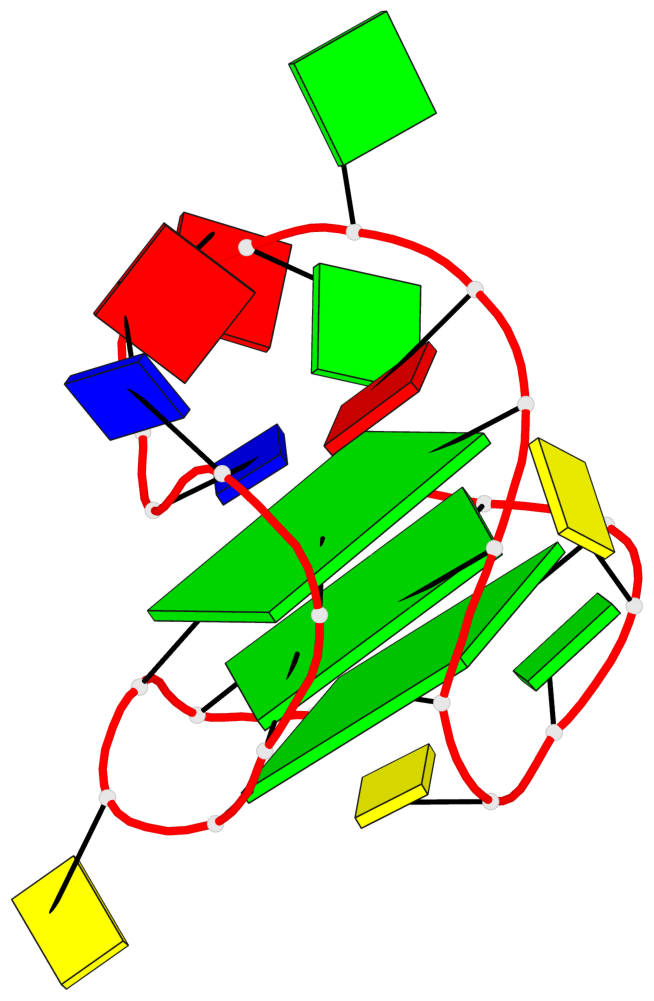
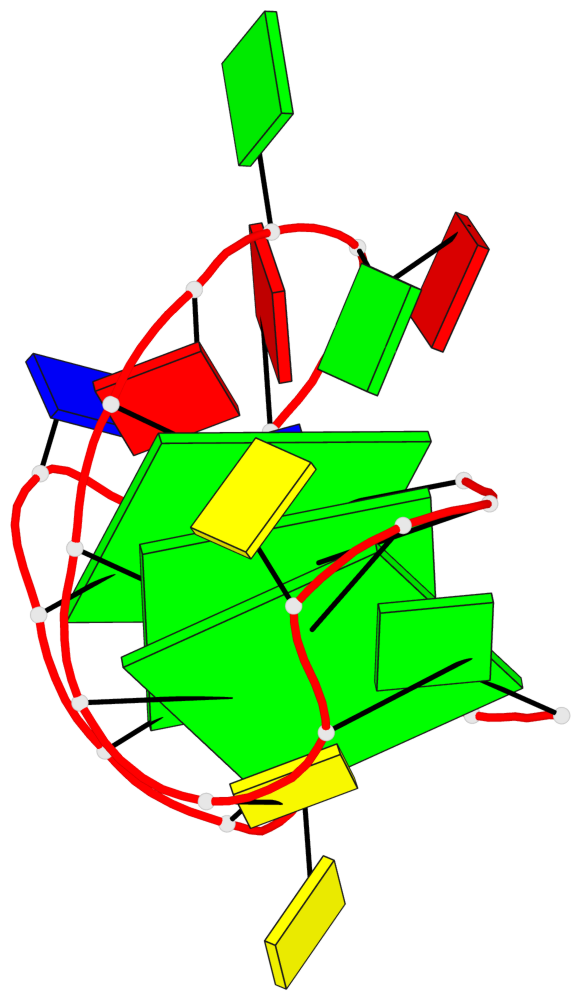
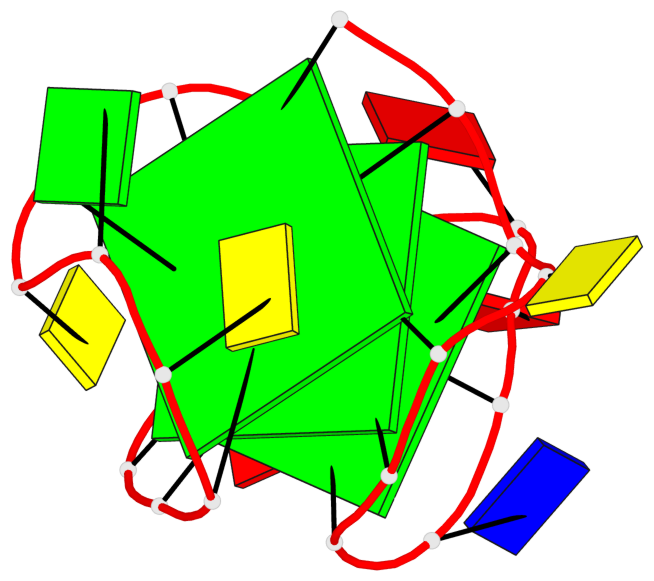
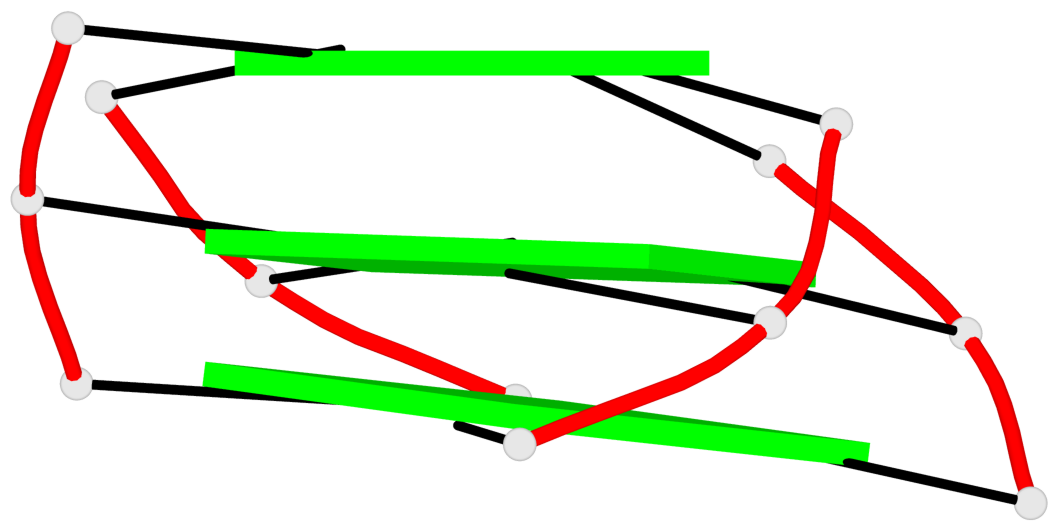
Base-block schematics in six views
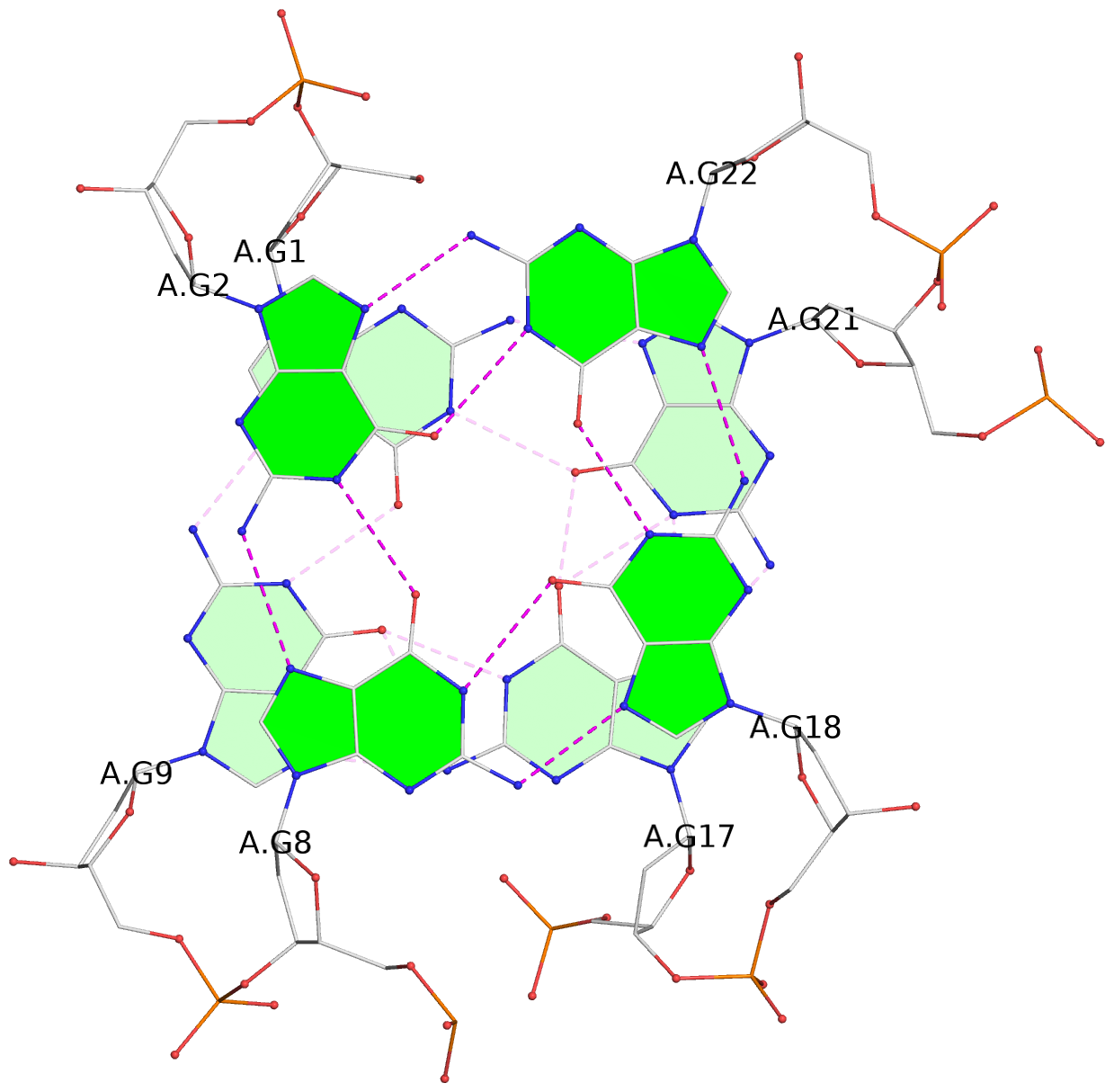
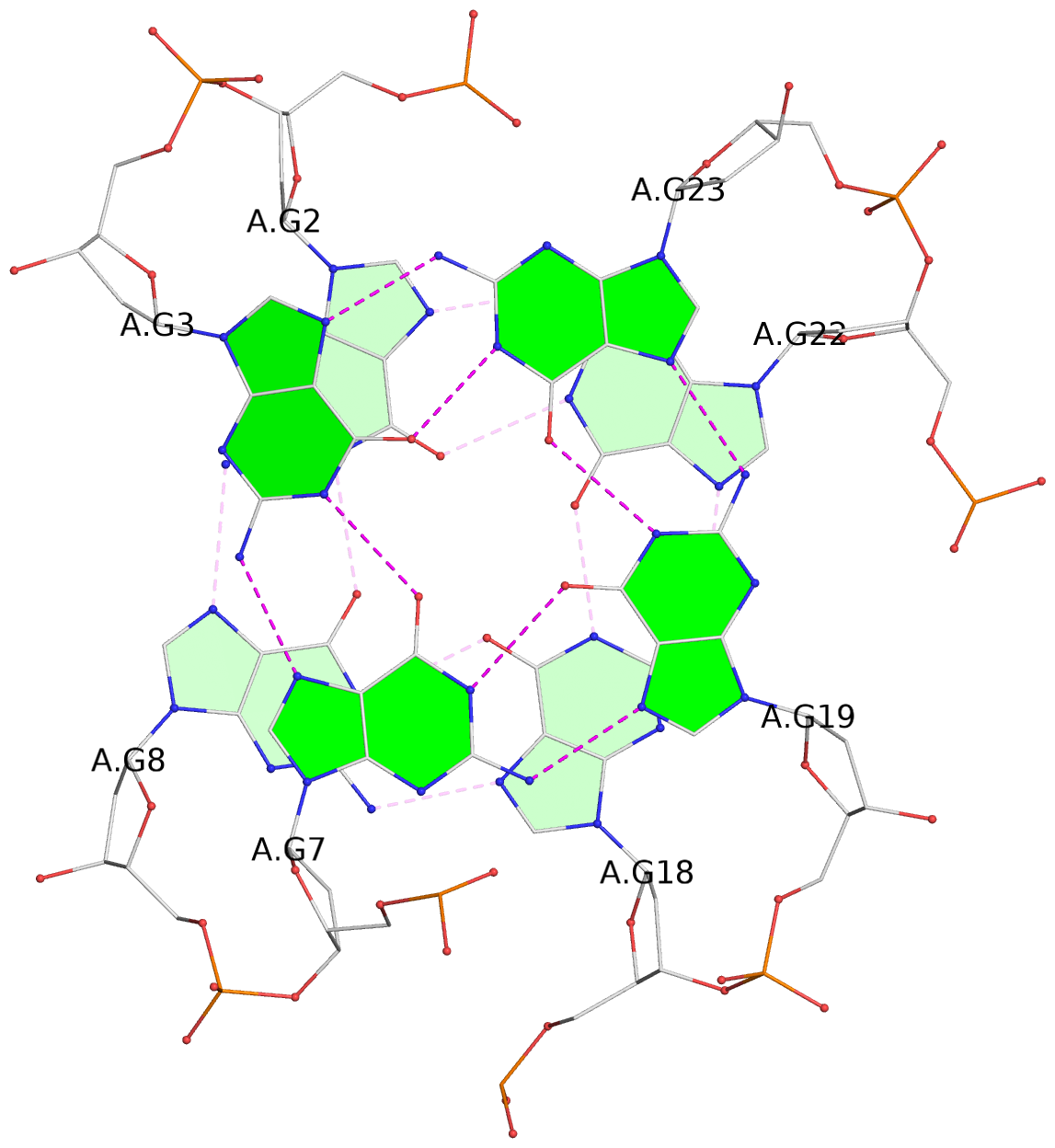
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=..-. groove=wn-- planarity=0.176 type=other nts=4 GGGG A.DG1,A.DG9,A.DG17,A.DG21 2 glyco-bond=-s-- sugar=.... groove=wn-- planarity=0.172 type=other nts=4 GGGG A.DG2,A.DG8,A.DG18,A.DG22 3 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.284 type=other nts=4 GGGG A.DG3,A.DG7,A.DG19,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.