Detailed DSSR results for the G-quadruplex: PDB entry 2idn
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2idn
- Class
- DNA
- Method
- NMR
- Summary
- NMR structure of a new modified thrombin binding aptamer containing a 5'-5' inversion of polarity site
- Reference
- Martino L, Virno A, Randazzo A, Virgilio A, Esposito V, Giancola C, Bucci M, Cirino G, Mayol L (2006): "A new modified thrombin binding aptamer containing a 5'-5' inversion of polarity site." Nucleic Acids Res., 34, 6653-6662. doi: 10.1093/nar/gkl915.
- Abstract
- The solution structure of a new modified thrombin binding aptamer (TBA) containing a 5'-5' inversion of polarity site, namely d(3'GGT5'-5'TGGTGTGGTTGG3'), is reported. NMR and CD spectroscopy, as well as molecular dynamic and mechanic calculations, have been used to characterize the 3D structure. The modified oligonucleotide is characterized by a chair-like structure consisting of two G-tetrads connected by three edge-wise TT, TGT and TT loops. d(3'GGT5'-5'TGGTGTGGTTGG3') is characterized by an unusual folding, being three strands parallel to each other and only one strand oriented in opposite manner. This led to an anti-anti-anti-syn and syn-syn-syn-anti arrangement of the Gs in the two tetrads. The thermal stability of the modified oligonucleotide is 4 degrees C higher than the corresponding unmodified TBA. d(3'GGT5'-5'TGGTGTGGTTGG3') continues to display an anticoagulant activity, even if decreased with respect to the TBA.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+Lm+Lw+Ln), UD3(1+3), DDUD
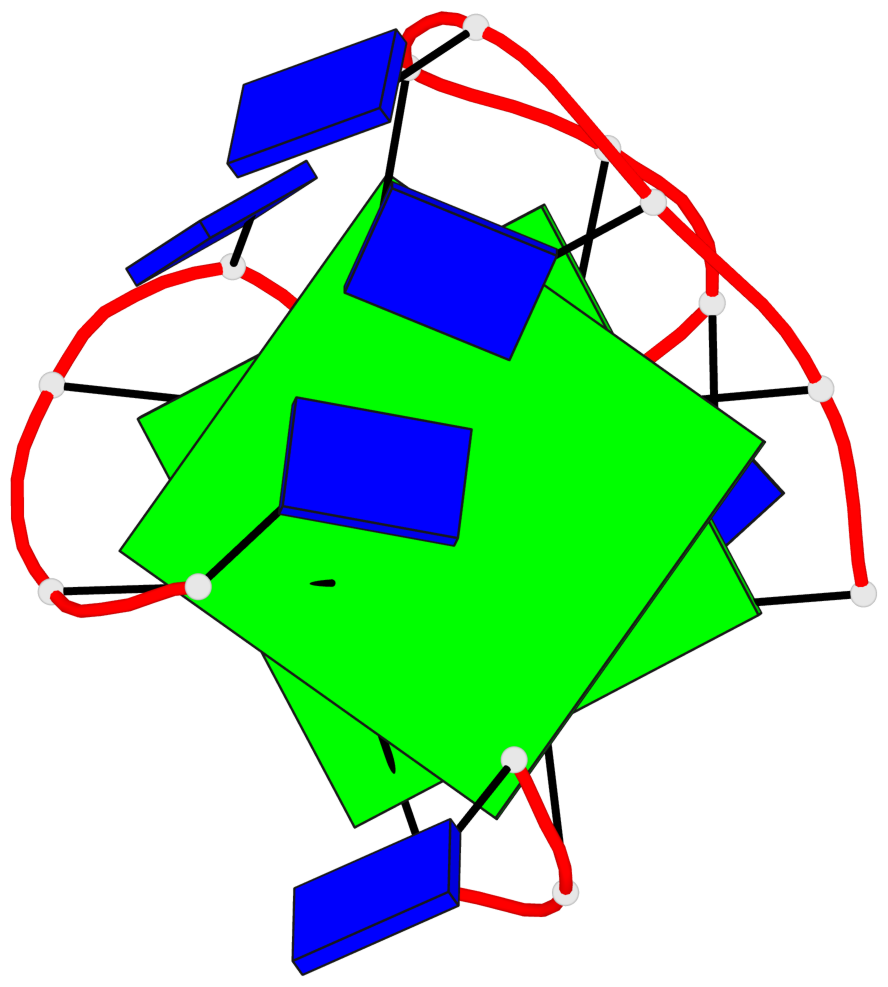
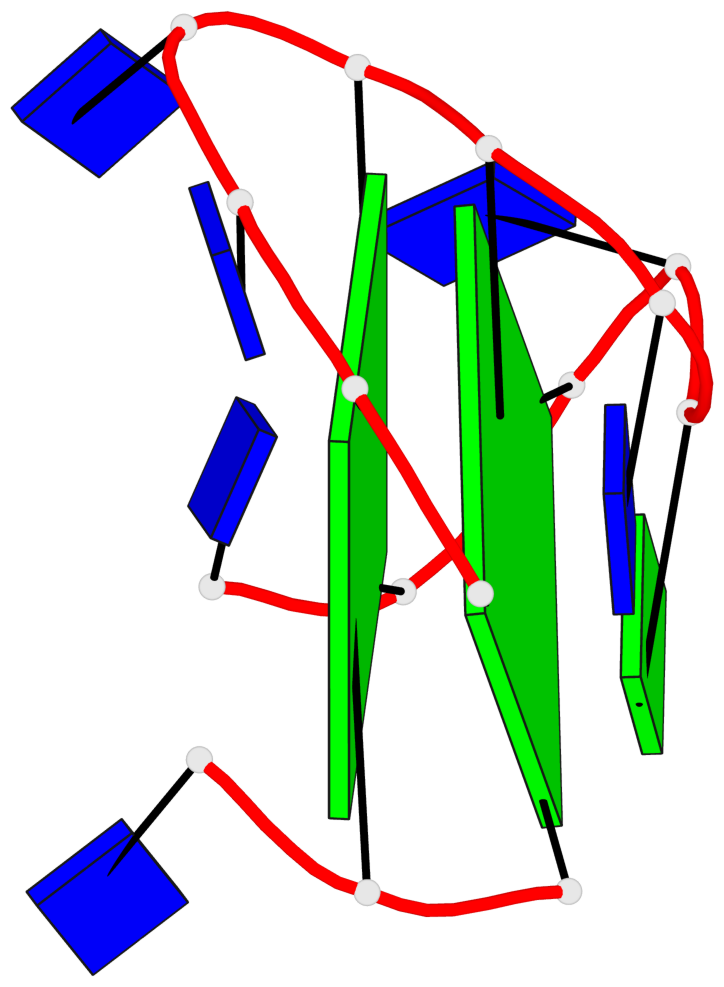
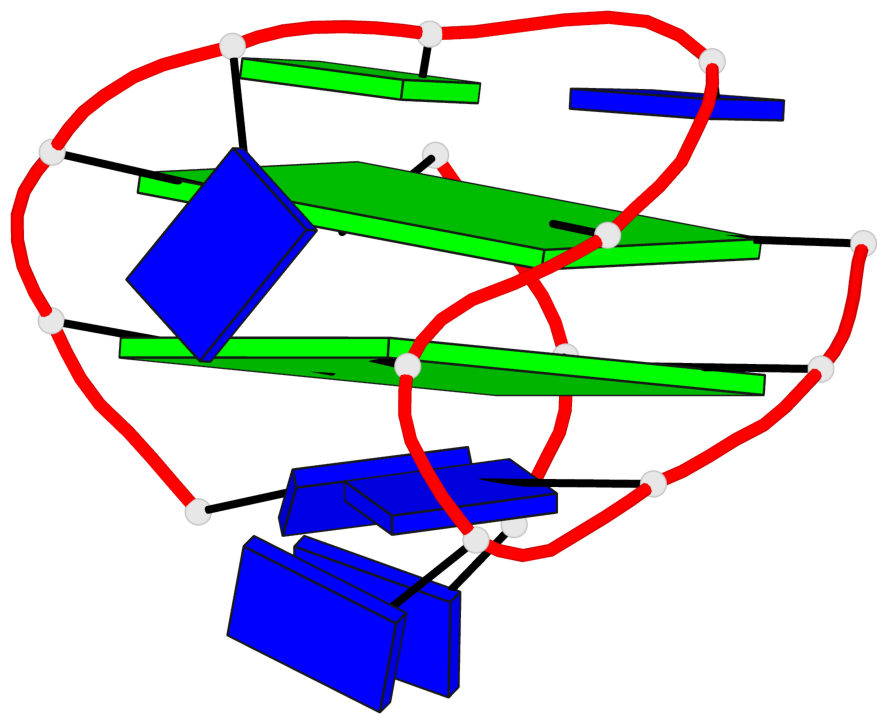
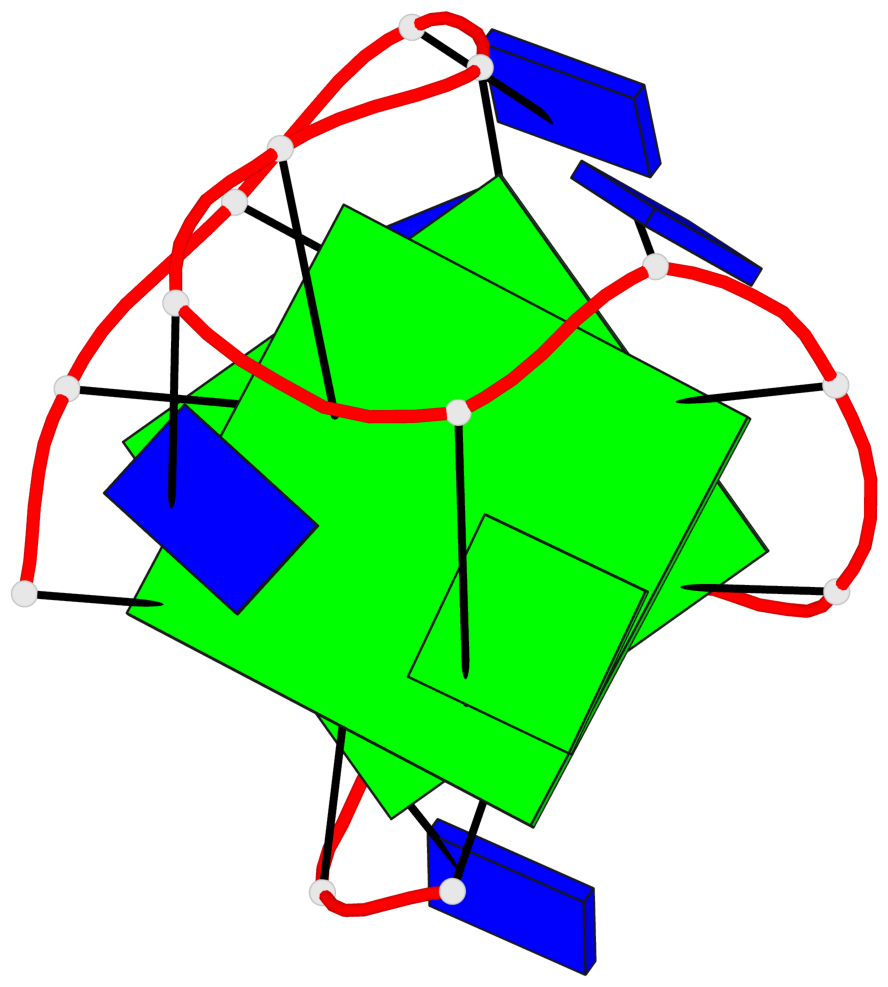
Base-block schematics in six views
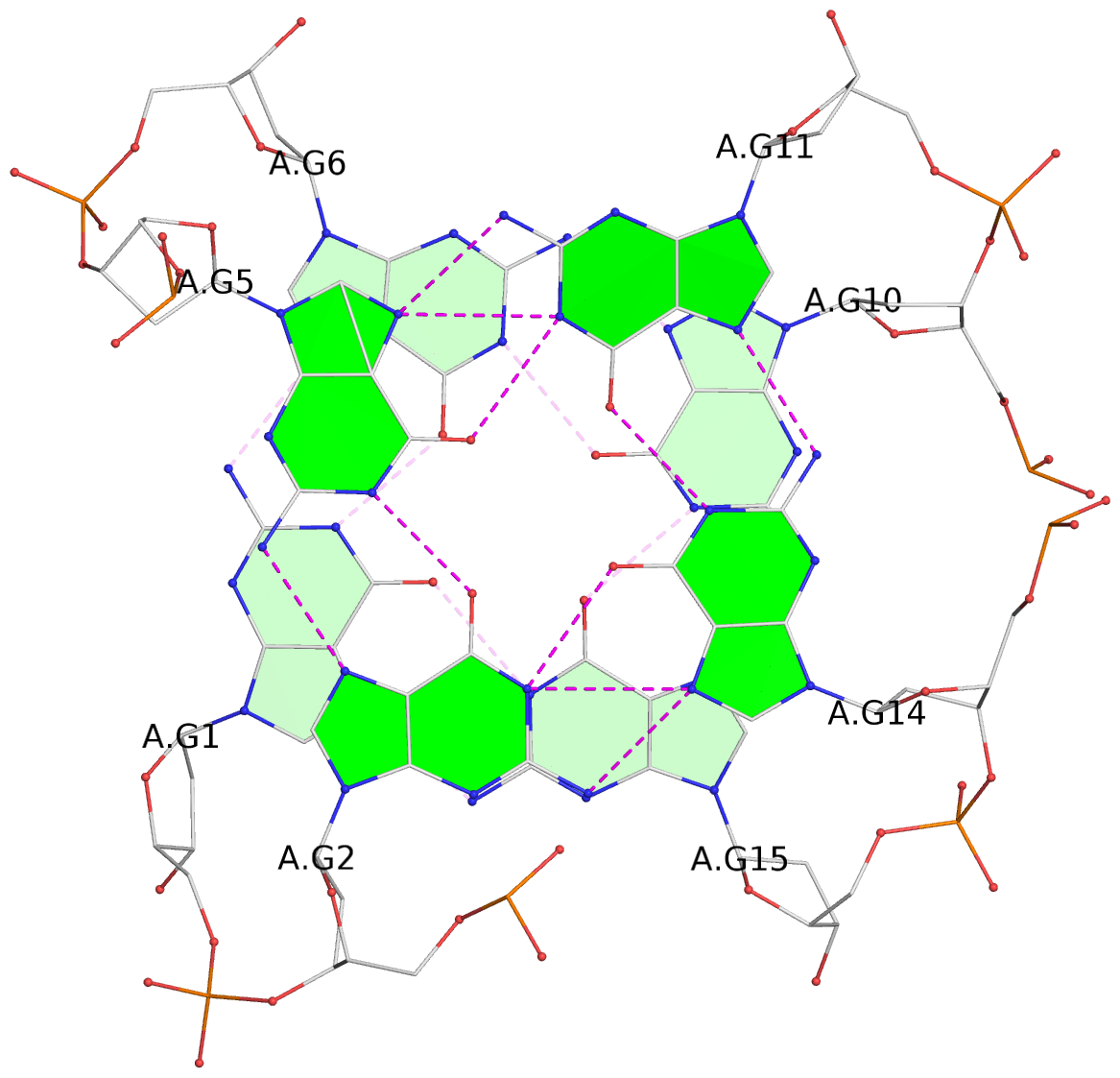
List of 2 G-tetrads
1 glyco-bond=--s- sugar=3--- groove=-wn- planarity=0.412 type=bowl nts=4 GGGG A.DG1,A.DG6,A.DG10,A.DG15 2 glyco-bond=ss-s sugar=---- groove=-wn- planarity=0.380 type=other nts=4 GGGG A.DG2,A.DG5,A.DG11,A.DG14
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.