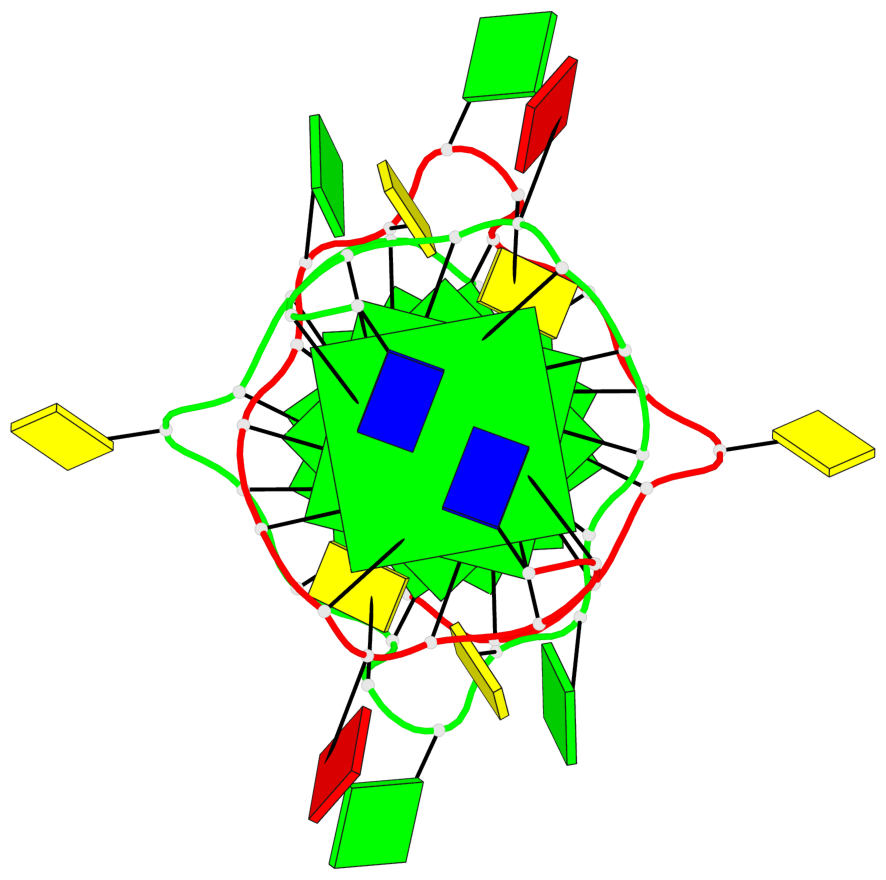
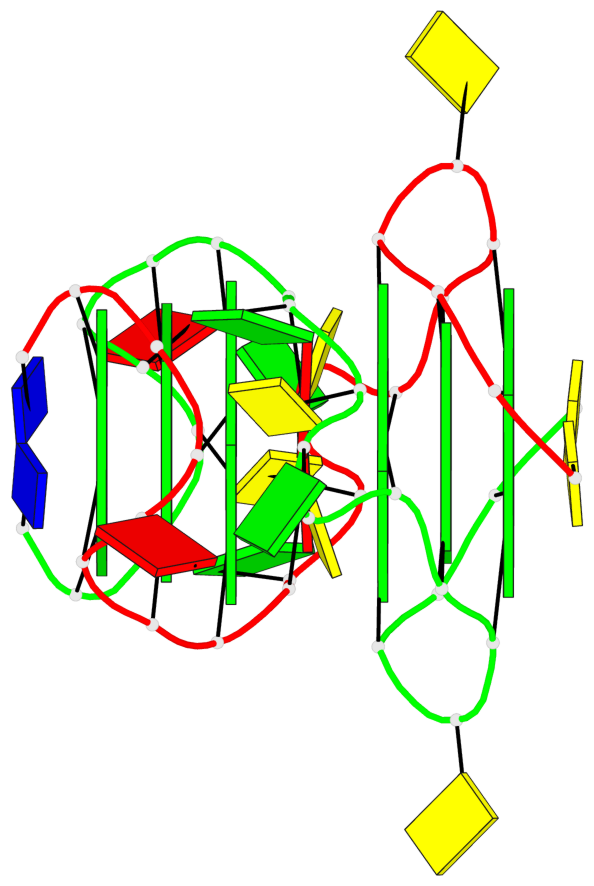
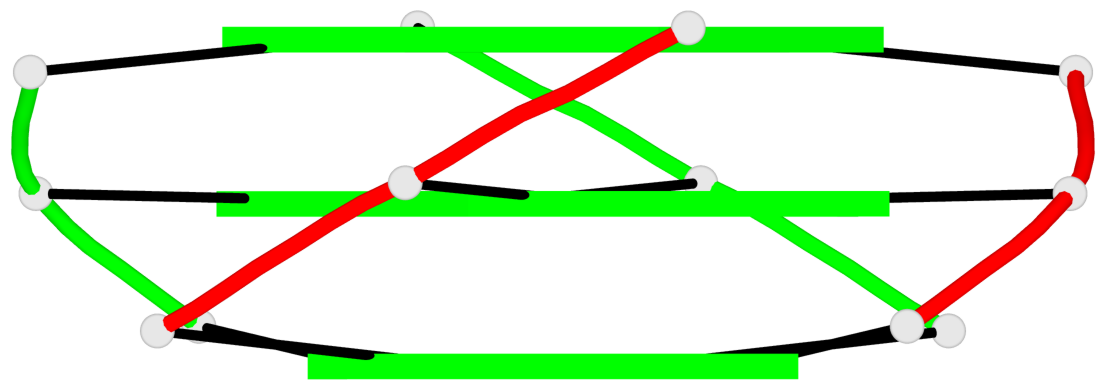
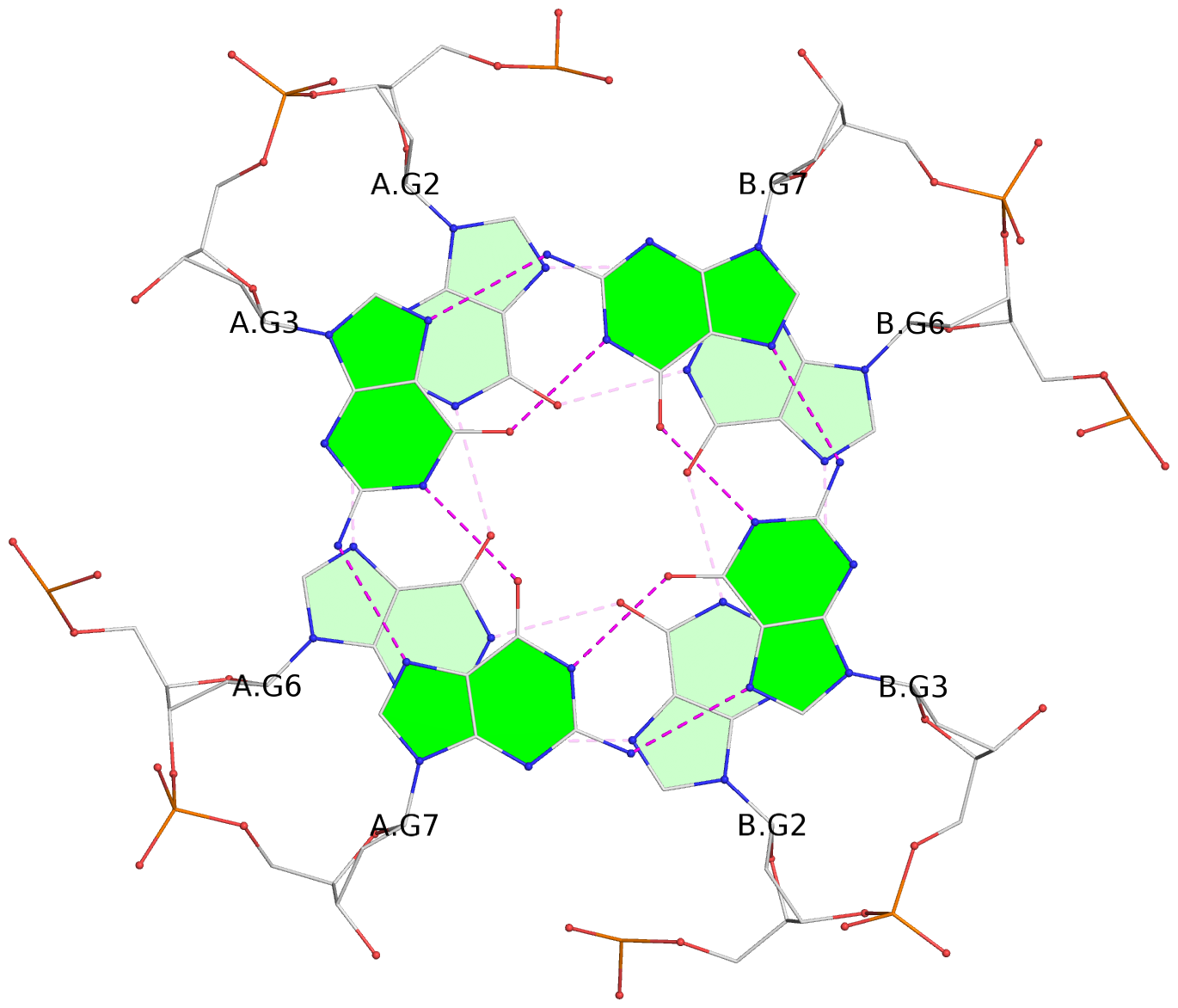
Detailed DSSR results for the G-quadruplex: PDB entry 2kyo
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2kyo
- Class
- DNA
- Method
- NMR
- Summary
- Dimeric human ckit-2 proto-oncogene promoter quadruplex DNA NMR, 10 structures
- Reference
- Kuryavyi V, Phan AT, Patel DJ (2010): "Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter." Nucleic Acids Res., 38, 6757-6773. doi: 10.1093/nar/gkq558.
- Abstract
- Previous studies have demonstrated that nuclease hypersensitivity regions of several proto-oncogenic DNA promoters, situated upstream of transcription start sites, contain guanine-rich tracts that form intramolecular G-quadruplexes stabilized by stacked G•G•G•G tetrads in monovalent cation solution. The human c-kit oncogenic promoter, an important target in the treatment of gastrointestinal tumors, contains two such stretches of guanine-rich tracts, designated c-kit1 and c-kit2. Our previous nuclear magnetic resonance (NMR)-based studies reported on the novel G-quadruplex scaffold of the c-kit1 promoter in K(+)-containing solution, where we showed for the first time that even an isolated guanine was involved in G-tetrad formation. These NMR-based studies are now extended to the c-kit2 promoter, which adopts two distinct all-parallel-stranded conformations in slow exchange, one of which forms a monomeric G-quadruplex (form-I) in 20 mM K(+)-containing solution and the other a novel dimeric G-quadruplex (form-II) in 100 mM K(+)-containing solution. The c-kit2 promoter dimeric form-II G-quadruplex adopts an unprecedented all-parallel-stranded topology where individual c-kit2 promoter strands span a pair of three-G-tetrad-layer-containing all-parallel-stranded G-quadruplexes aligned in a 3' to 5'-end orientation, with stacking continuity between G-quadruplexes mediated by a sandwiched A•A non-canonical pair. We propose that strand exchange during recombination events within guanine-rich segments, could potentially be mediated by a synapsis intermediate involving an intergenic parallel-stranded dimeric G-quadruplex.
- G4 notes
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, parallel(4+0), UUUU
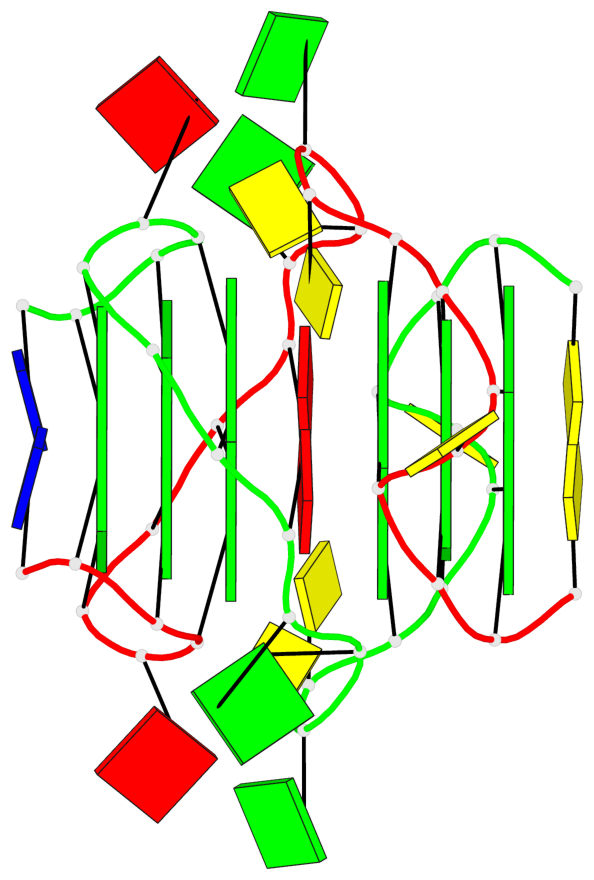
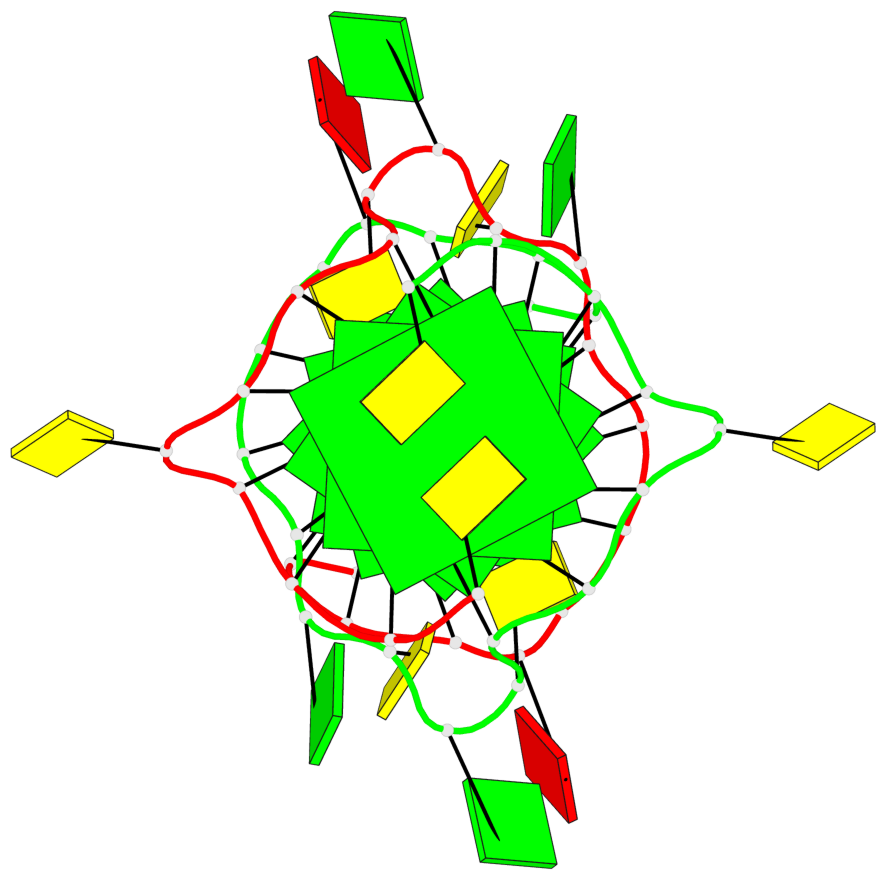
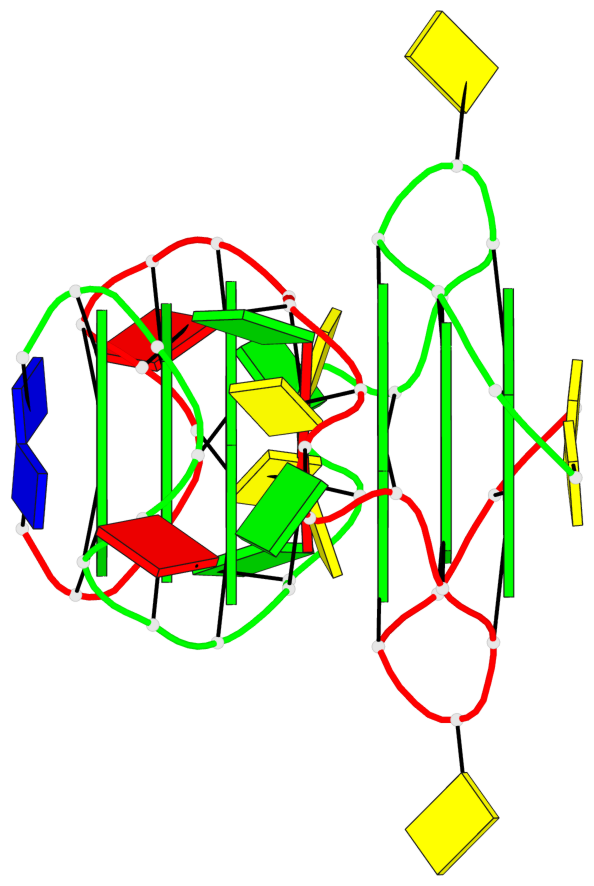
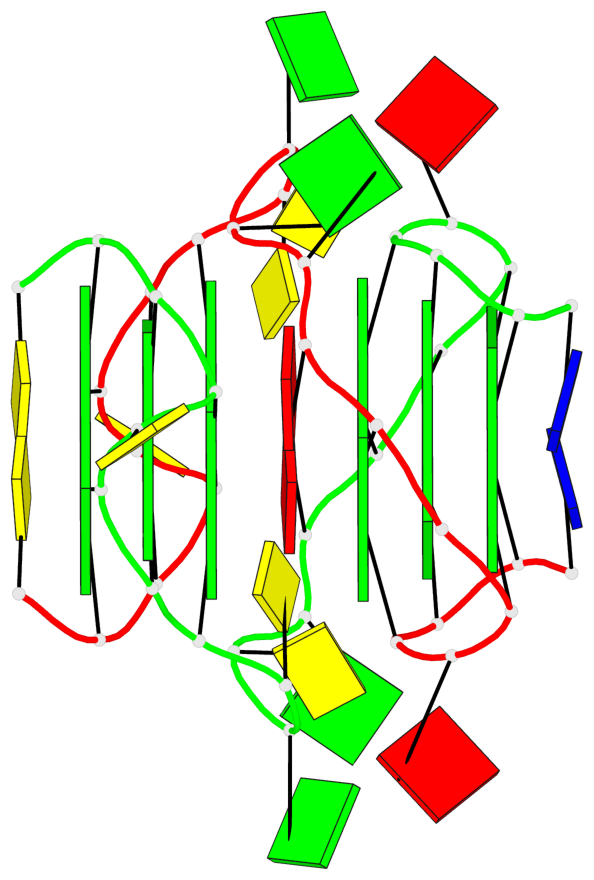
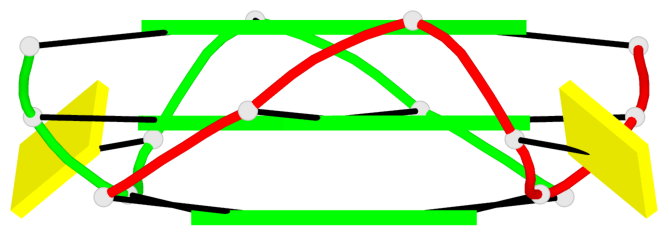
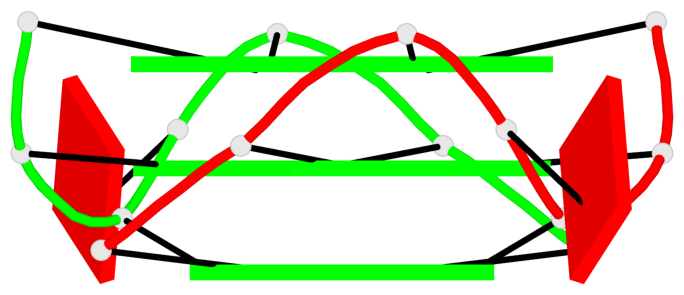
Base-block schematics in six views
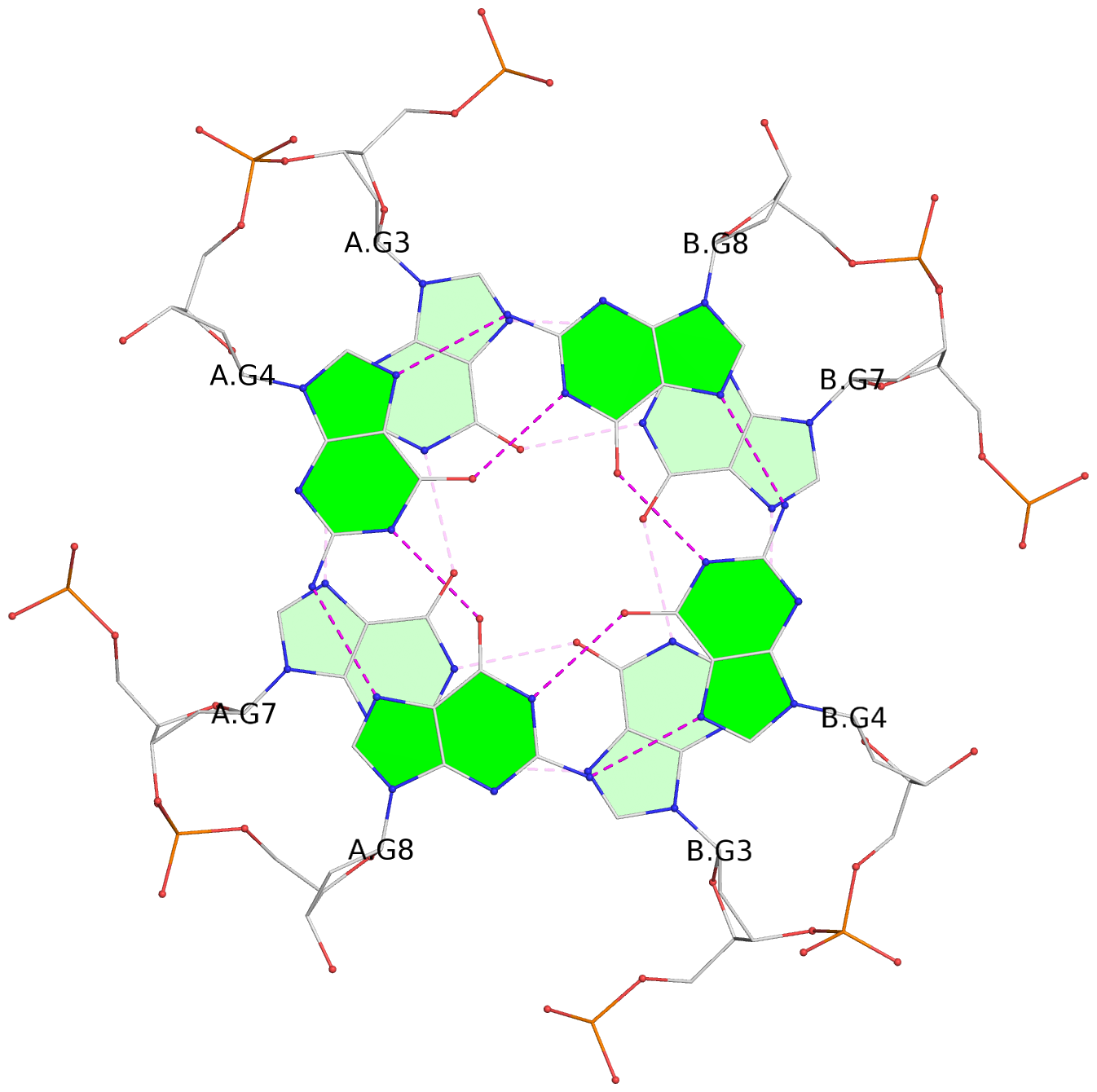
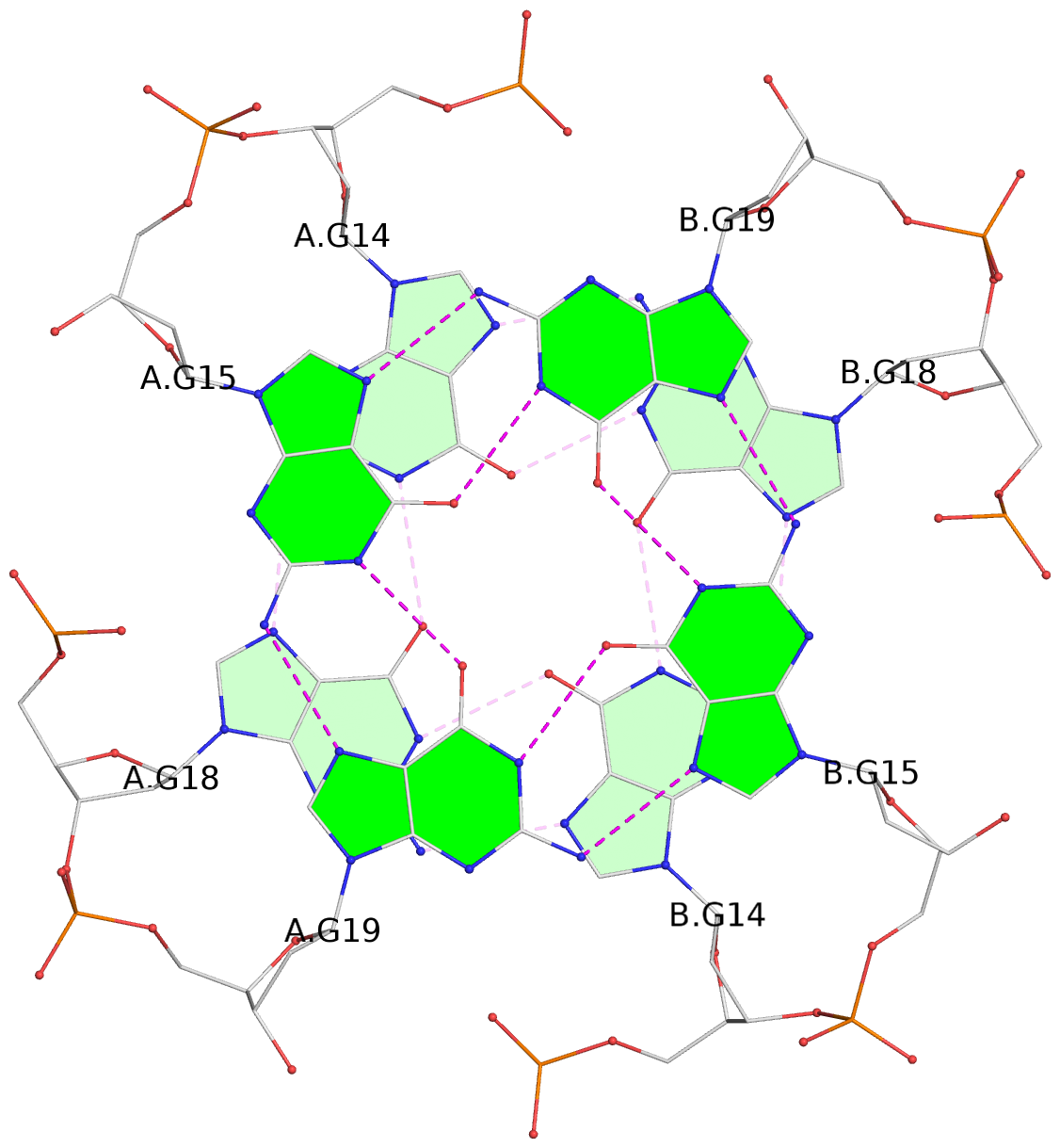
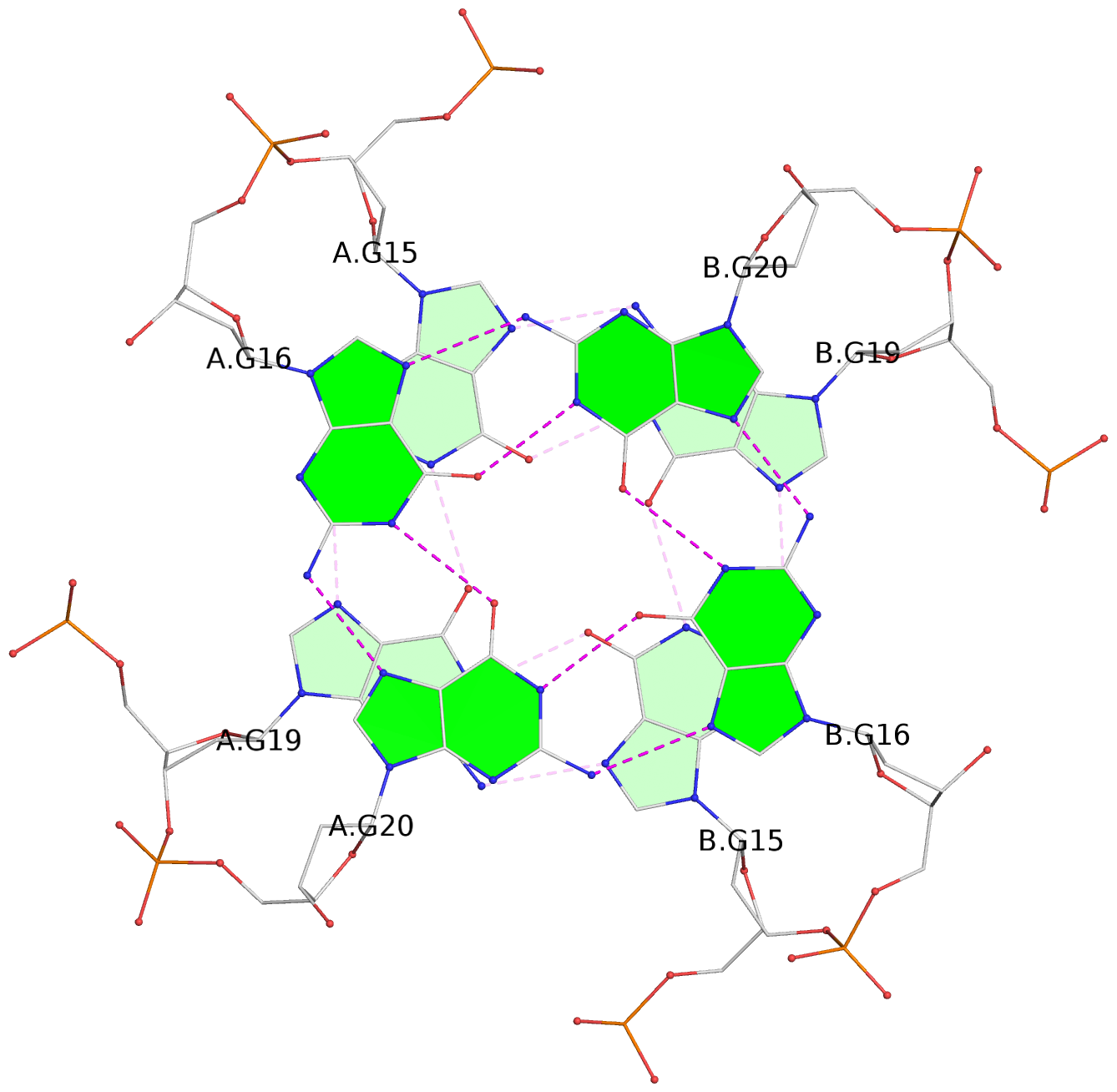
List of 6 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.044 type=planar nts=4 GGGG A.DG2,A.DG6,B.DG2,B.DG6 2 glyco-bond=---- sugar=---- groove=---- planarity=0.065 type=planar nts=4 GGGG A.DG3,A.DG7,B.DG3,B.DG7 3 glyco-bond=---- sugar=---- groove=---- planarity=0.140 type=planar nts=4 GGGG A.DG4,A.DG8,B.DG4,B.DG8 4 glyco-bond=---- sugar=---- groove=---- planarity=0.106 type=planar nts=4 GGGG A.DG14,A.DG18,B.DG14,B.DG18 5 glyco-bond=---- sugar=---- groove=---- planarity=0.051 type=planar nts=4 GGGG A.DG15,A.DG19,B.DG15,B.DG19 6 glyco-bond=---- sugar=---- groove=---- planarity=0.179 type=other nts=4 GGGG A.DG16,A.DG20,B.DG16,B.DG20
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.