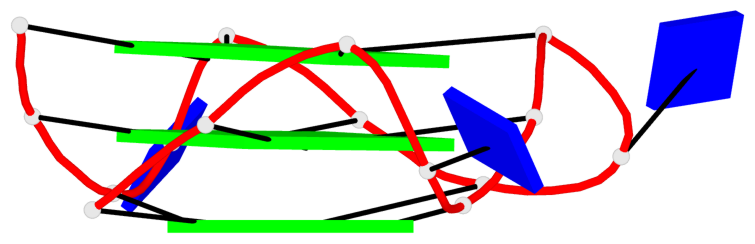
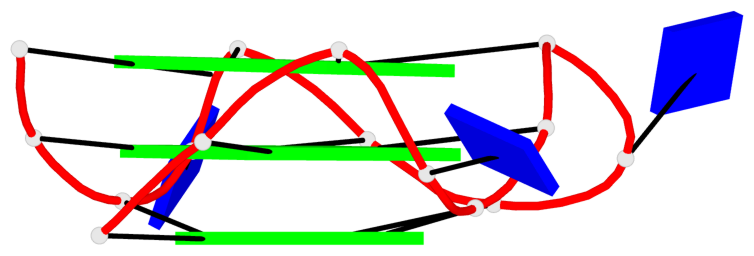
Detailed DSSR results for the G-quadruplex: PDB entry 2le6
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2le6
- Class
- DNA
- Method
- NMR
- Summary
- Structure of a dimeric all-parallel-stranded g-quadruplex stacked via the 5'-to-5' interface
- Reference
- Do NQ, Lim KW, Teo MH, Heddi B, Phan AT (2011): "Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity." Nucleic Acids Res. doi: 10.1093/nar/gkr539.
- Abstract
- G-rich oligonucleotides T30695 (or T30923), with the sequence of (GGGT)(4), and T40214, with the sequence of (GGGC)(4), have been reported to exhibit anti-HIV and anticancer activity. Here we report on the structure of a dimeric G-quadruplex adopted by a derivative of these sequences in K(+) solution. It comprises two identical propeller-type parallel-stranded G-quadruplex subunits each containing three G-tetrad layers that are stacked via the 5'-5' interface. We demonstrated control over the stacking of the two monomeric subunits by sequence modifications. Our analysis of possible structures at the stacking interface provides a general principle for stacking of G-quadruplexes, which could have implications for the assembly and recognition of higher-order G-quadruplex structures.
- G4 notes
- 6 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 3(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
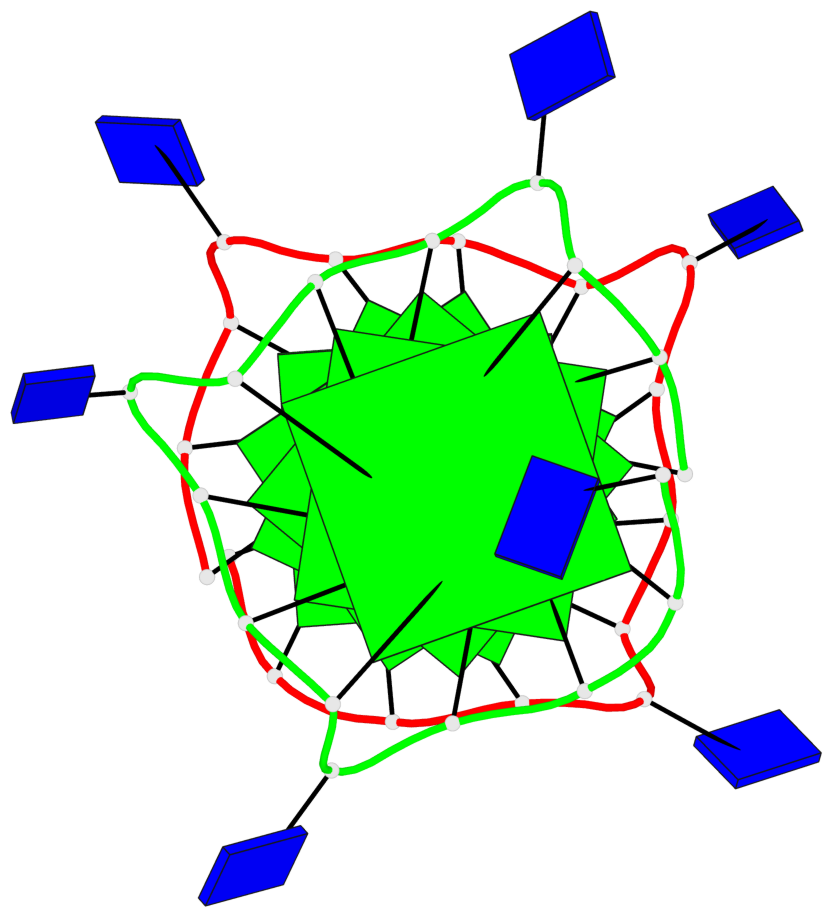
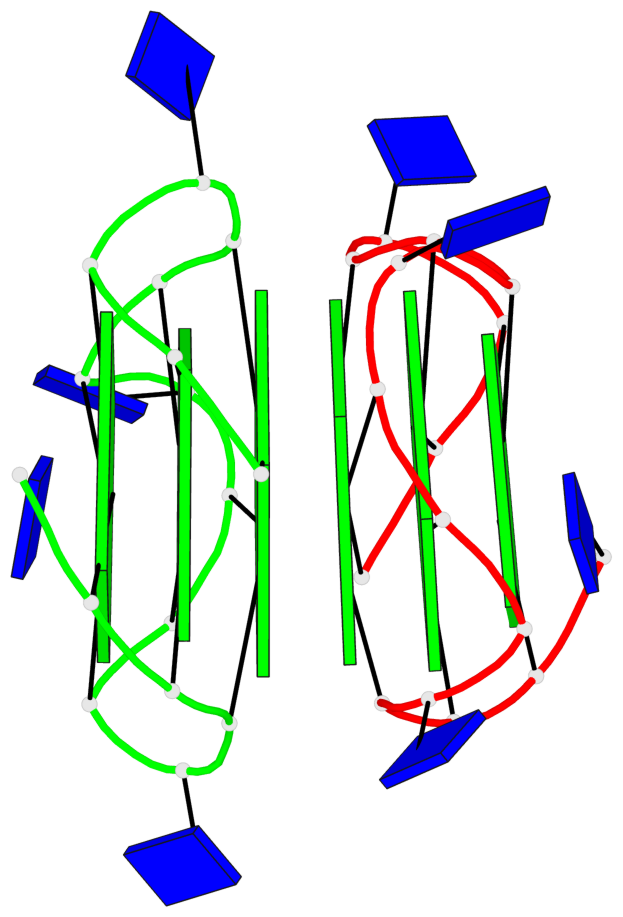
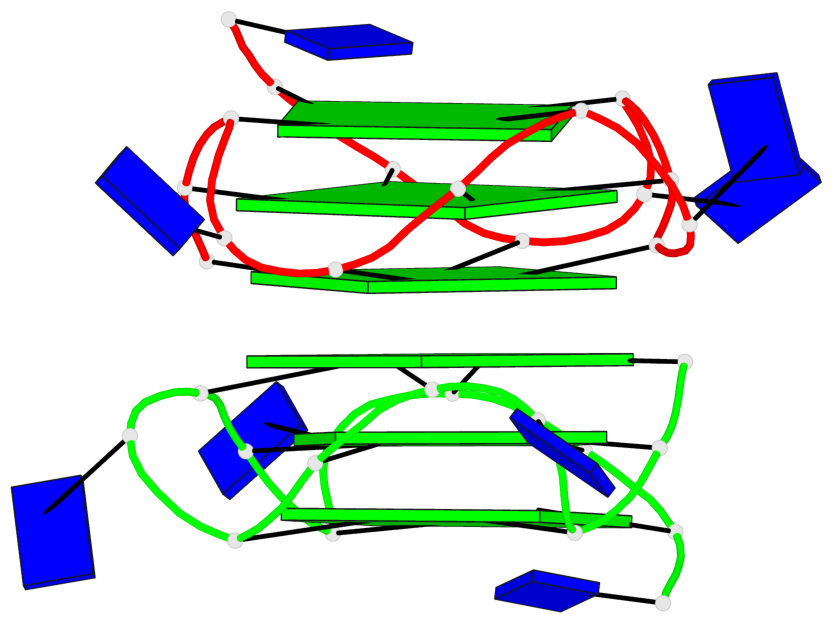
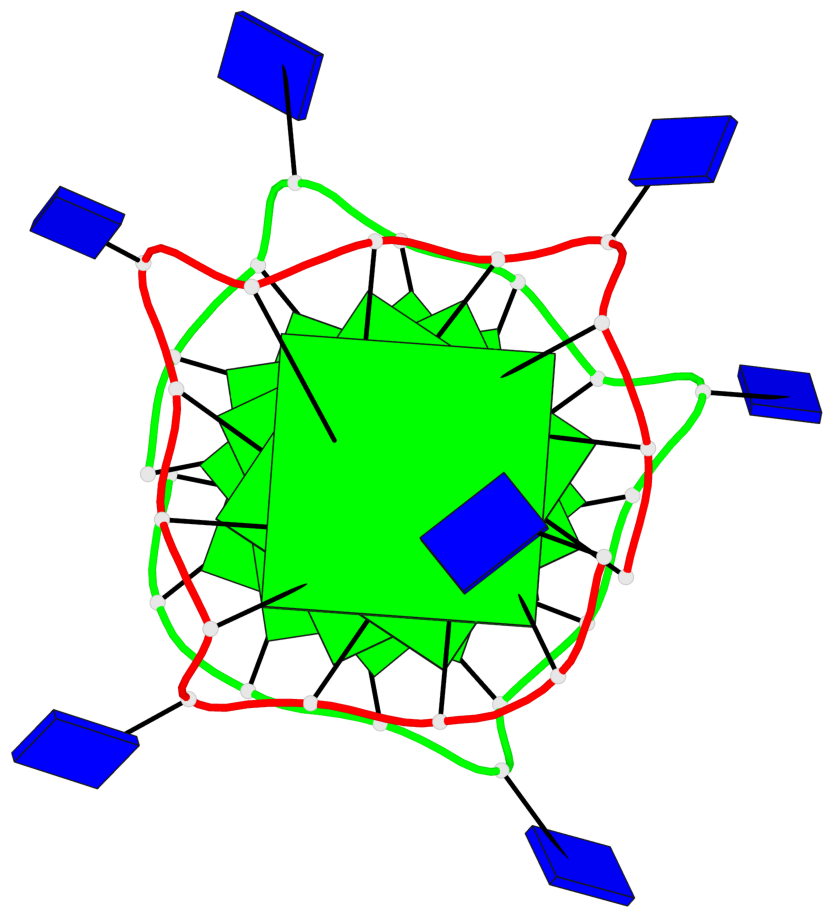
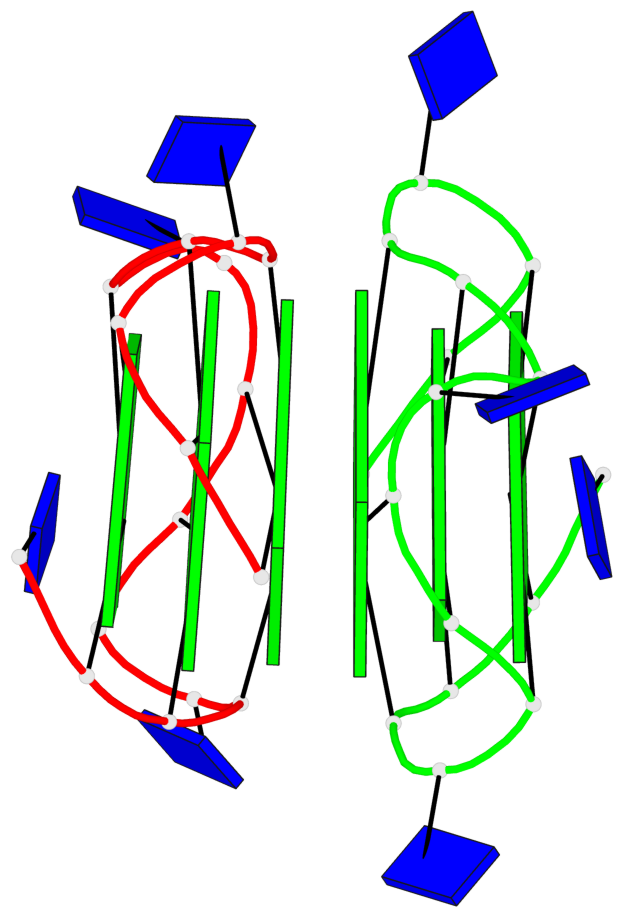
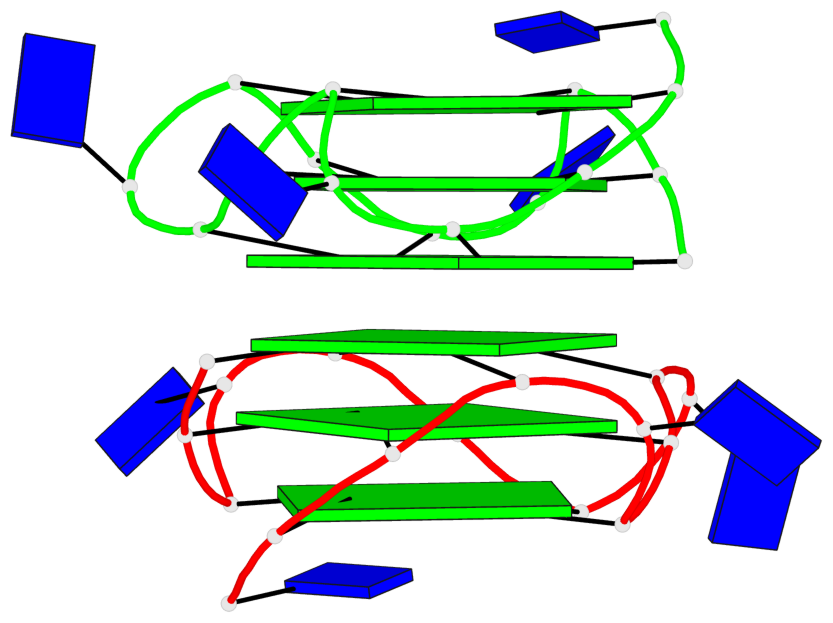
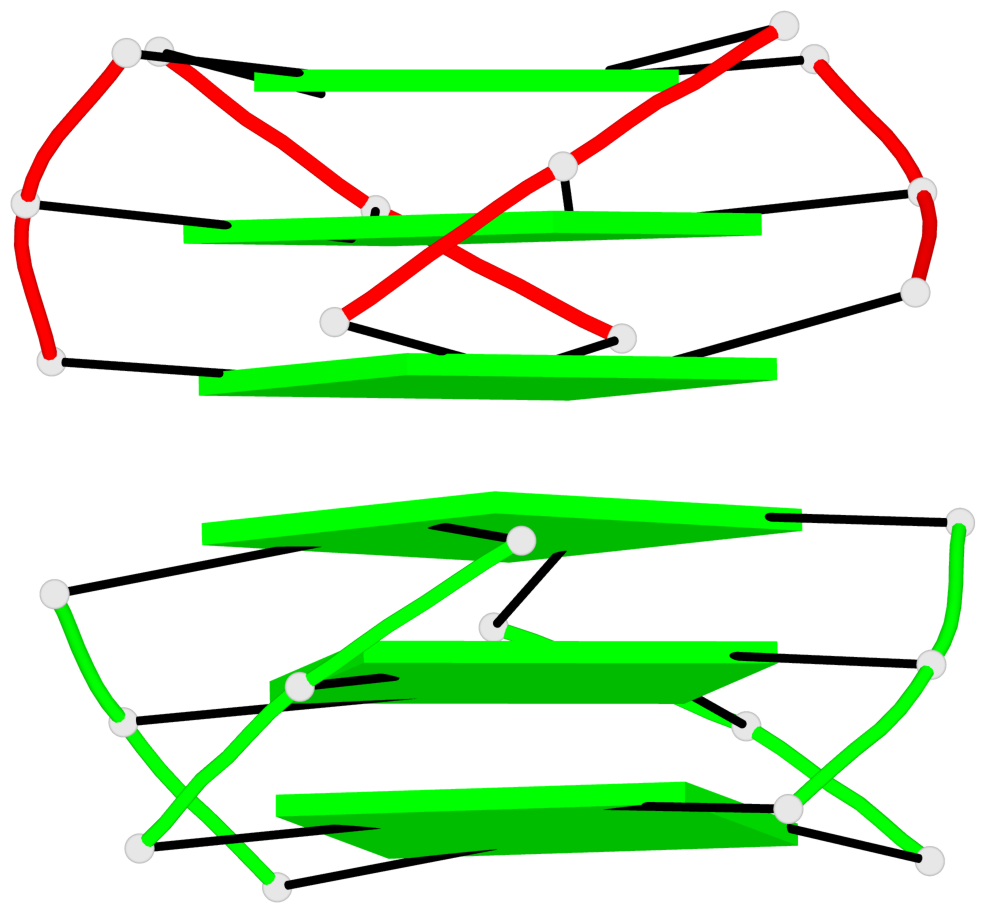
Base-block schematics in six views
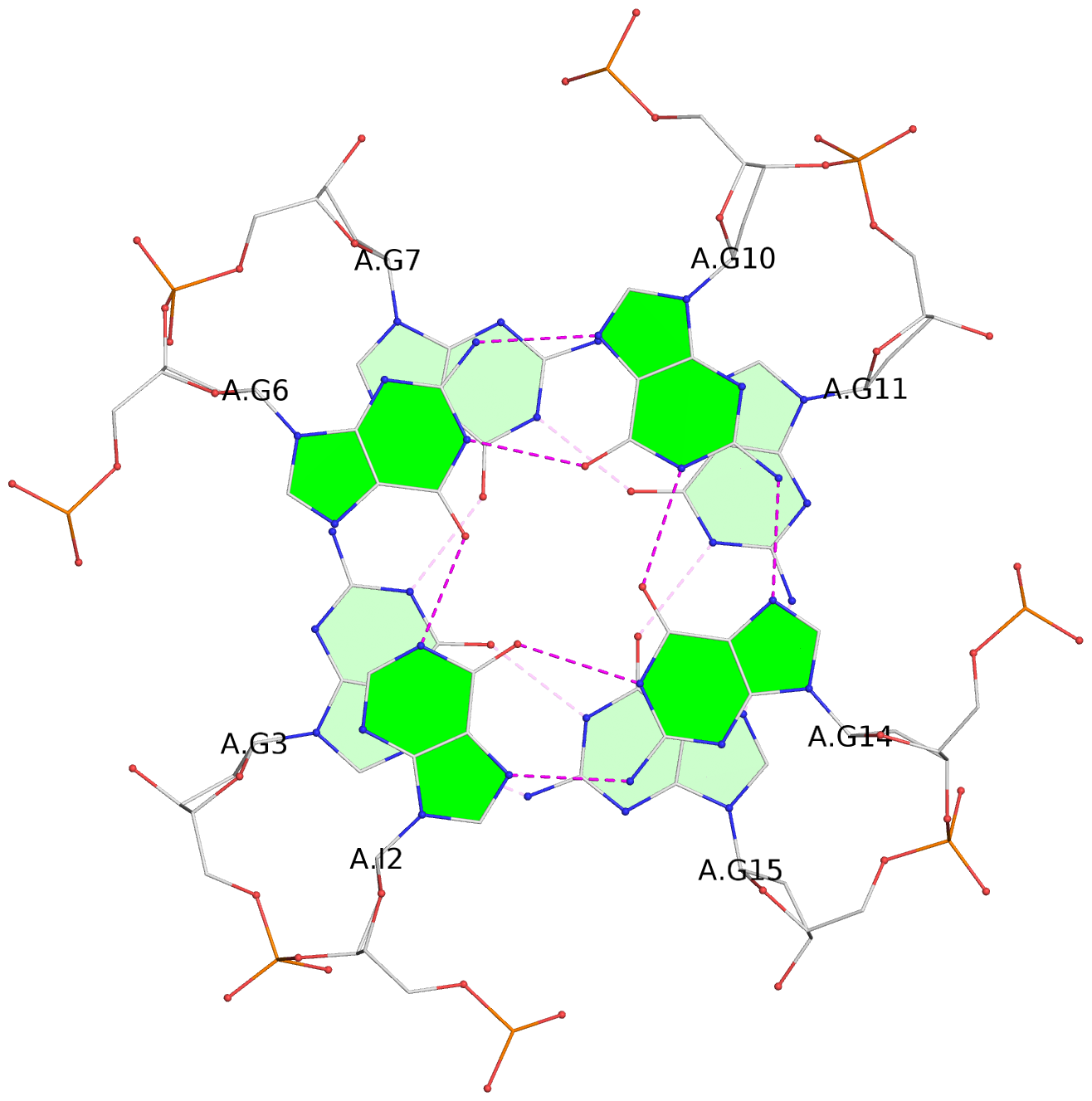
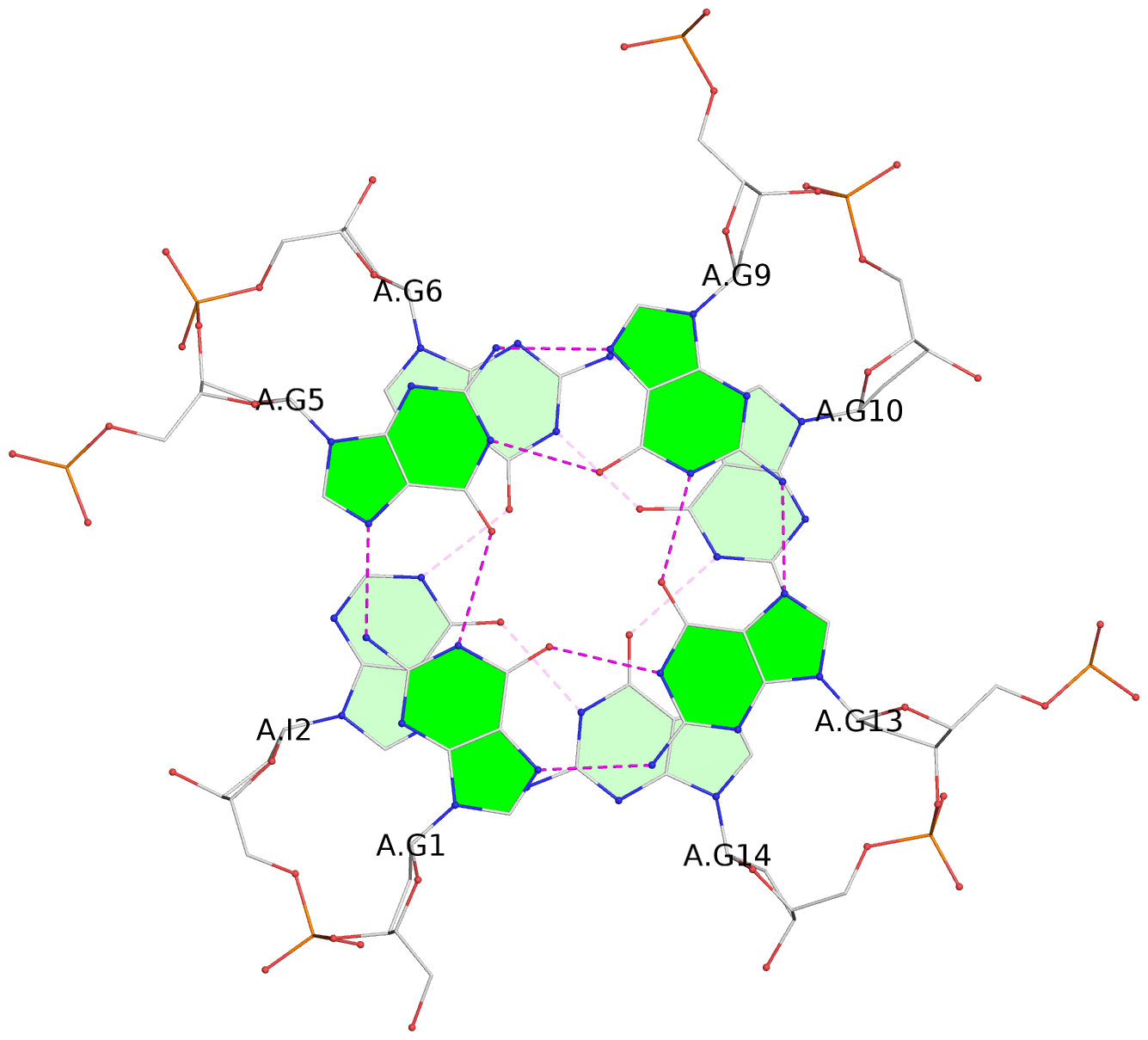
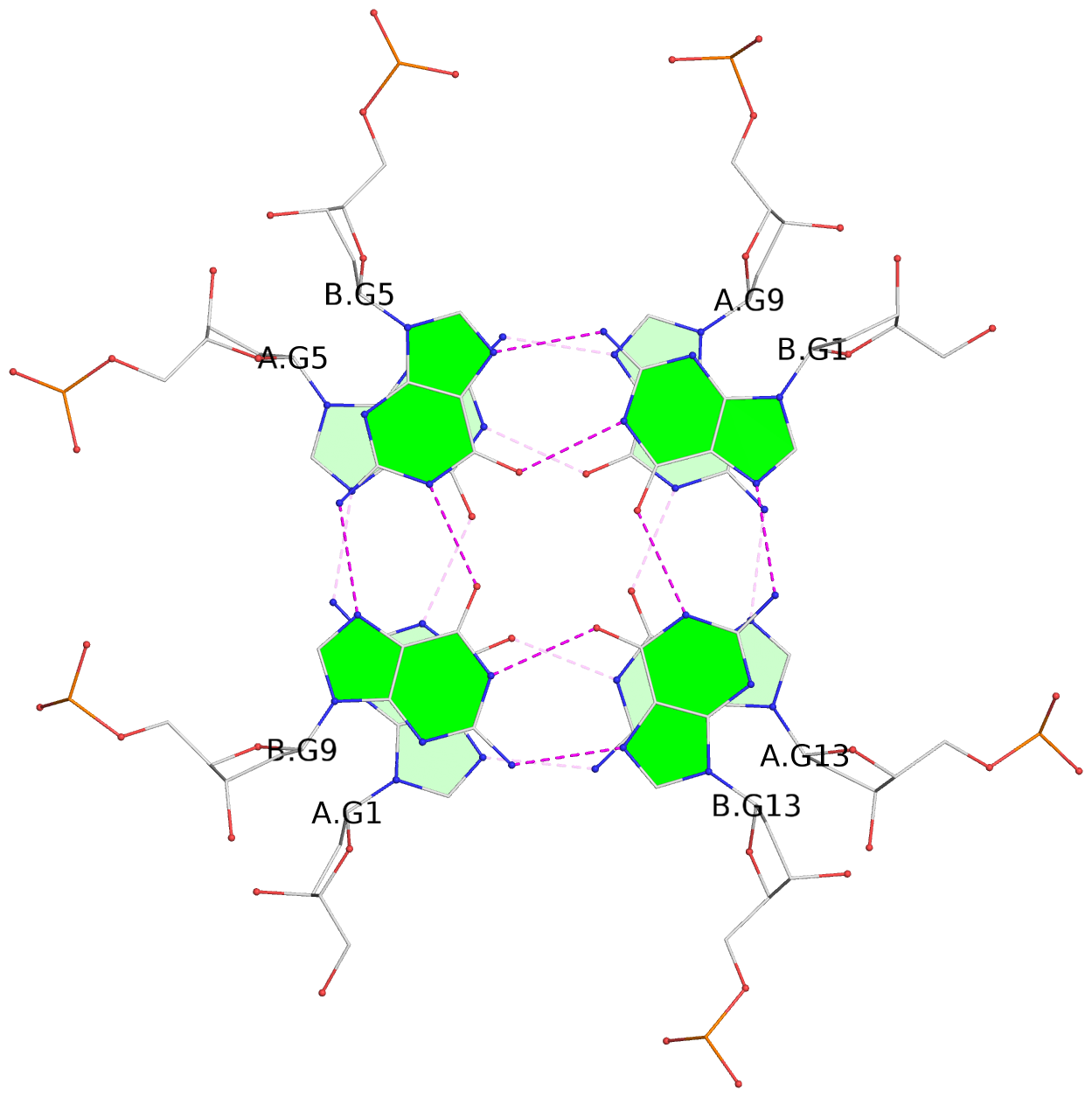
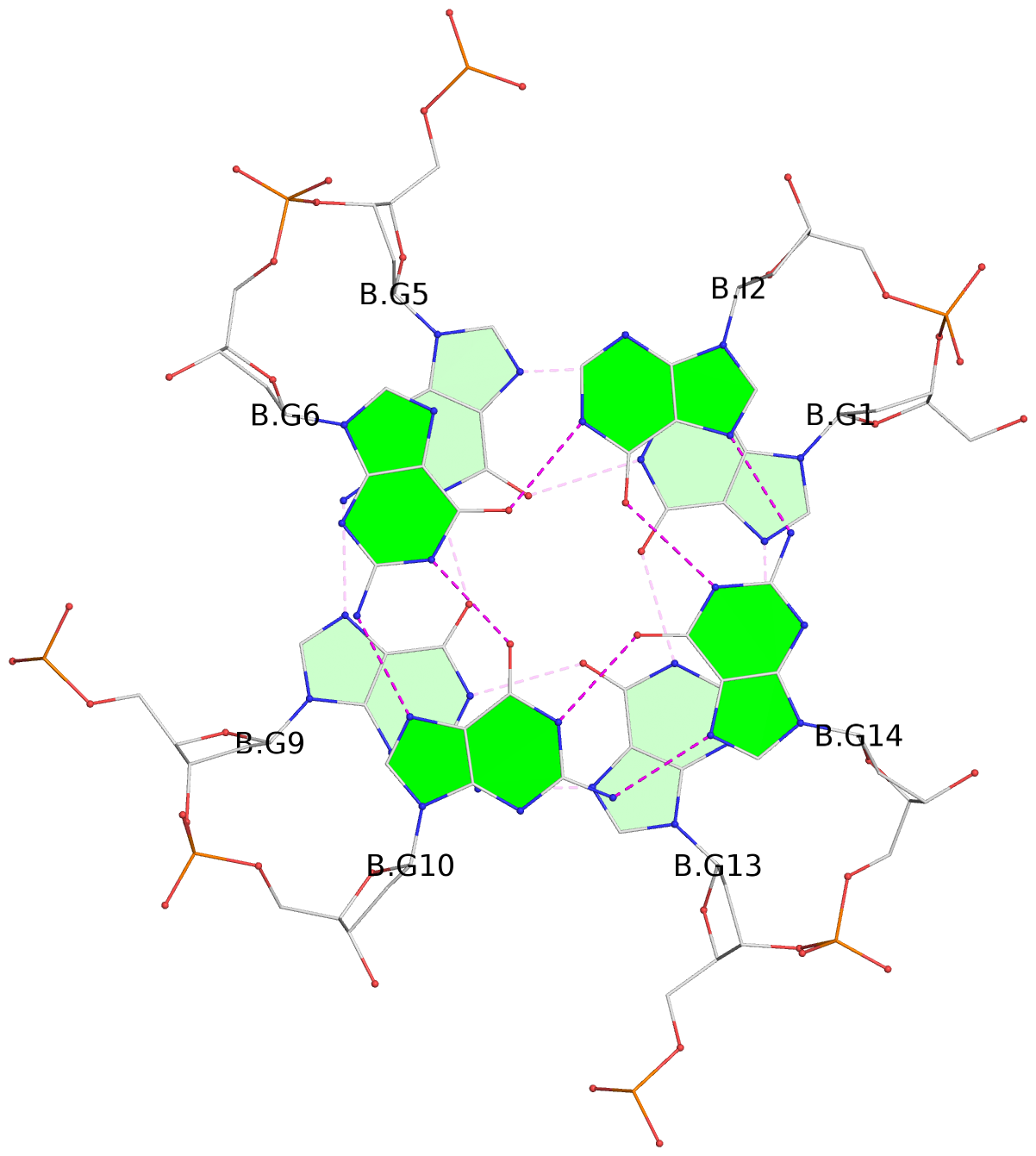
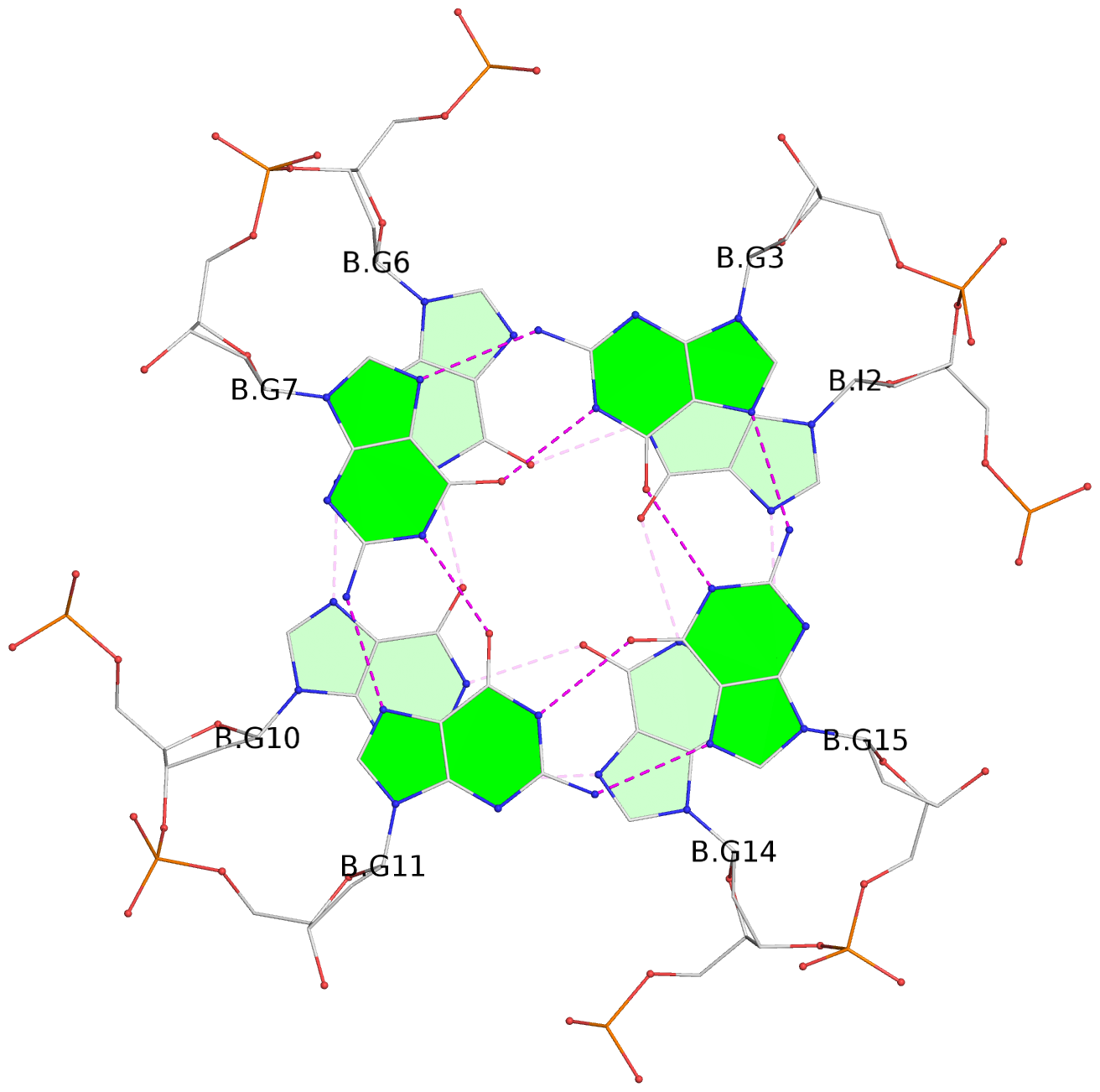
List of 6 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.130 type=planar nts=4 GGGG A.DG1,A.DG5,A.DG9,A.DG13 2 glyco-bond=---- sugar=---- groove=---- planarity=0.118 type=planar nts=4 gGGG A.DI2,A.DG6,A.DG10,A.DG14 3 glyco-bond=---- sugar=--.- groove=---- planarity=0.170 type=other nts=4 GGGG A.DG3,A.DG7,A.DG11,A.DG15 4 glyco-bond=---- sugar=---- groove=---- planarity=0.208 type=other nts=4 GGGG B.DG1,B.DG5,B.DG9,B.DG13 5 glyco-bond=---- sugar=---- groove=---- planarity=0.186 type=other nts=4 gGGG B.DI2,B.DG6,B.DG10,B.DG14 6 glyco-bond=---- sugar=--.- groove=---- planarity=0.170 type=other nts=4 GGGG B.DG3,B.DG7,B.DG11,B.DG15
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#2, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']