Detailed DSSR results for the G-quadruplex: PDB entry 3ibk
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 3ibk
- Class
- RNA
- Method
- X-ray (2.2 Å)
- Summary
- Crystal structure of a telomeric RNA quadruplex
- Reference
- Collie GW, Haider SM, Neidle S, Parkinson GN (2010): "A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex." Nucleic Acids Res., 38, 5569-5580. doi: 10.1093/nar/gkq259.
- Abstract
- DNA telomeric repeats in mammalian cells are transcribed to guanine-rich RNA sequences, which adopt parallel-stranded G-quadruplexes with a propeller-like fold. The successful crystallization and structure analysis of a bimolecular human telomeric RNA G-quadruplex, folded into the same crystalline environment as an equivalent DNA oligonucleotide sequence, is reported here. The structural basis of the increased stability of RNA telomeric quadruplexes over DNA ones and their preference for parallel topologies is described here. Our findings suggest that the 2'-OH hydroxyl groups in the RNA quadruplex play a significant role in redefining hydration structure in the grooves and the hydrogen bonding networks. The preference for specific nucleotides to populate the C3'-endo sugar pucker domain is accommodated by alterations in the phosphate backbone, which leads to greater stability through enhanced hydrogen bonding networks. Molecular dynamics simulations on the DNA and RNA quadruplexes are consistent with these findings. The computations, based on the native crystal structure, provide an explanation for RNA G-quadruplex ligand binding selectivity for a group of naphthalene diimide ligands as compared to the DNA G-quadruplex.
- G4 notes
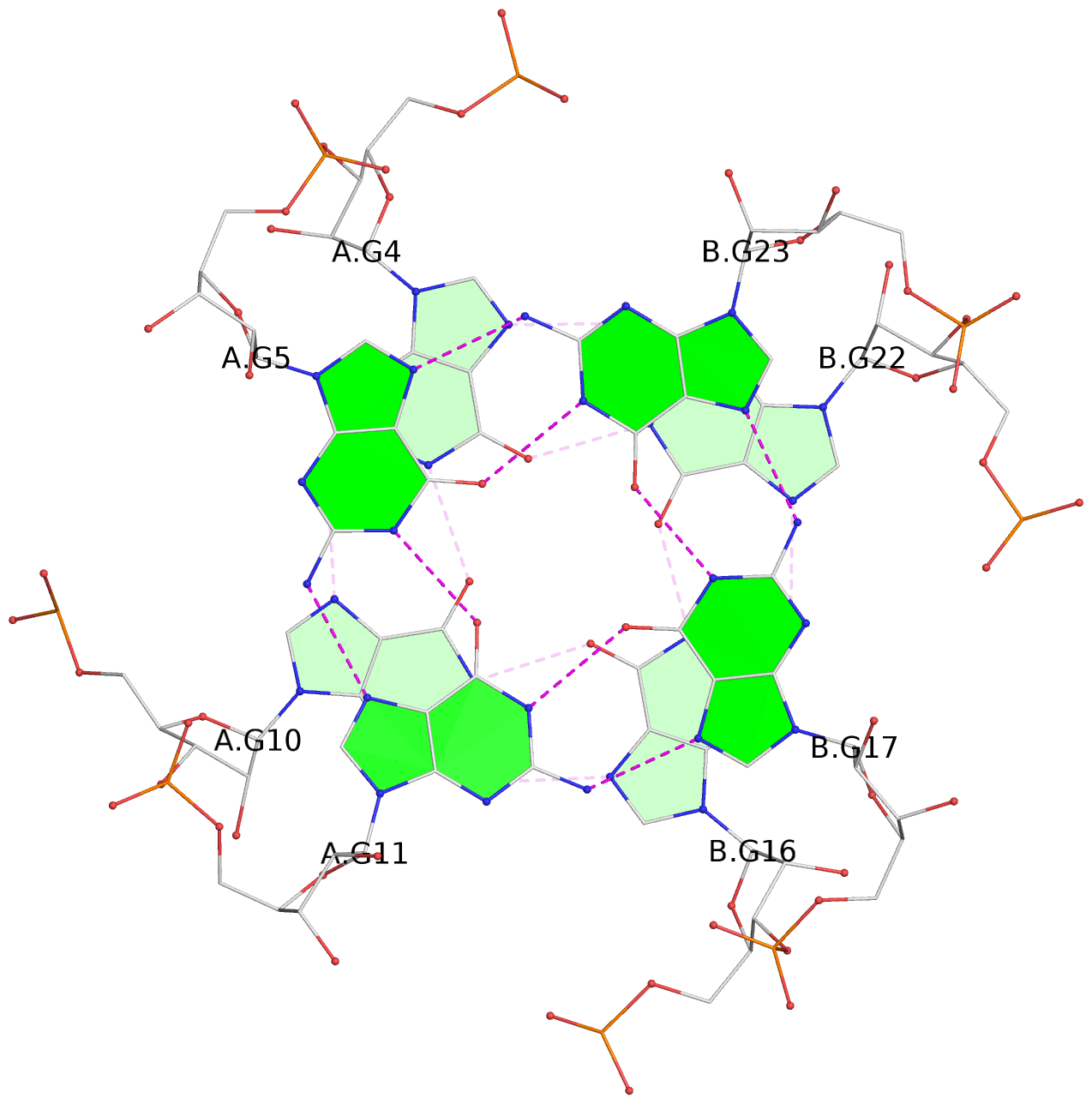
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
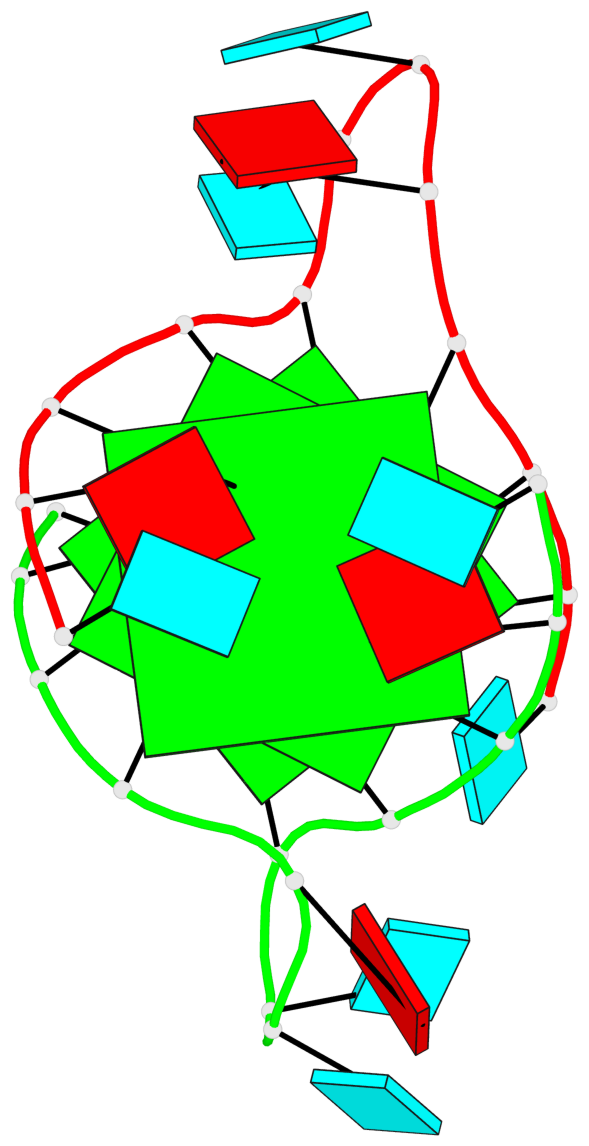
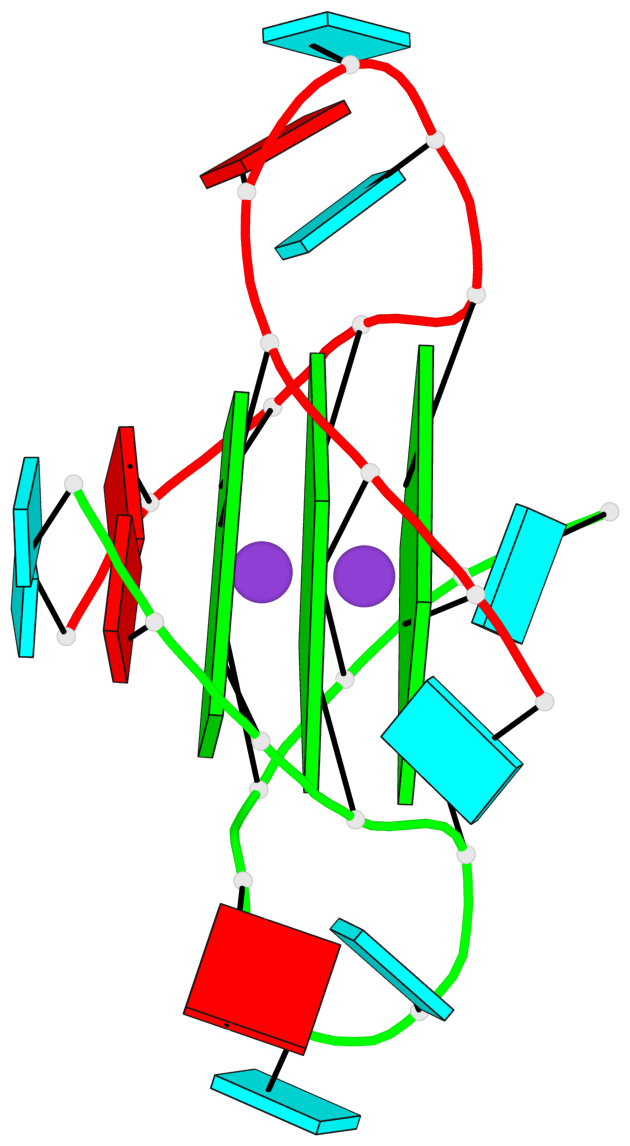
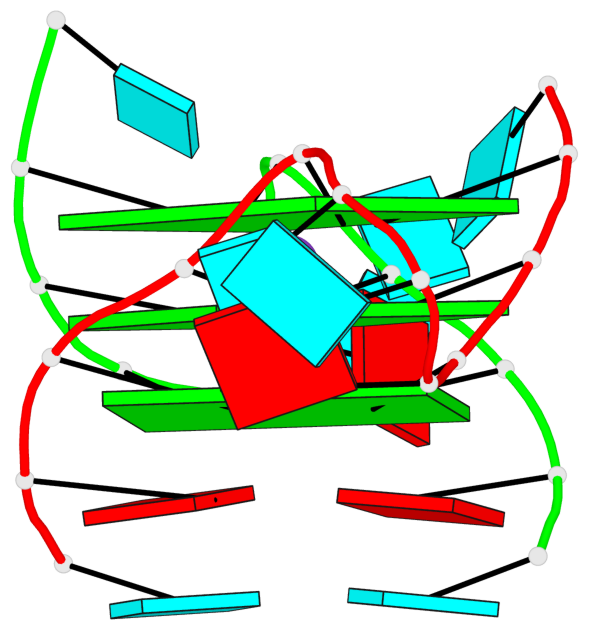
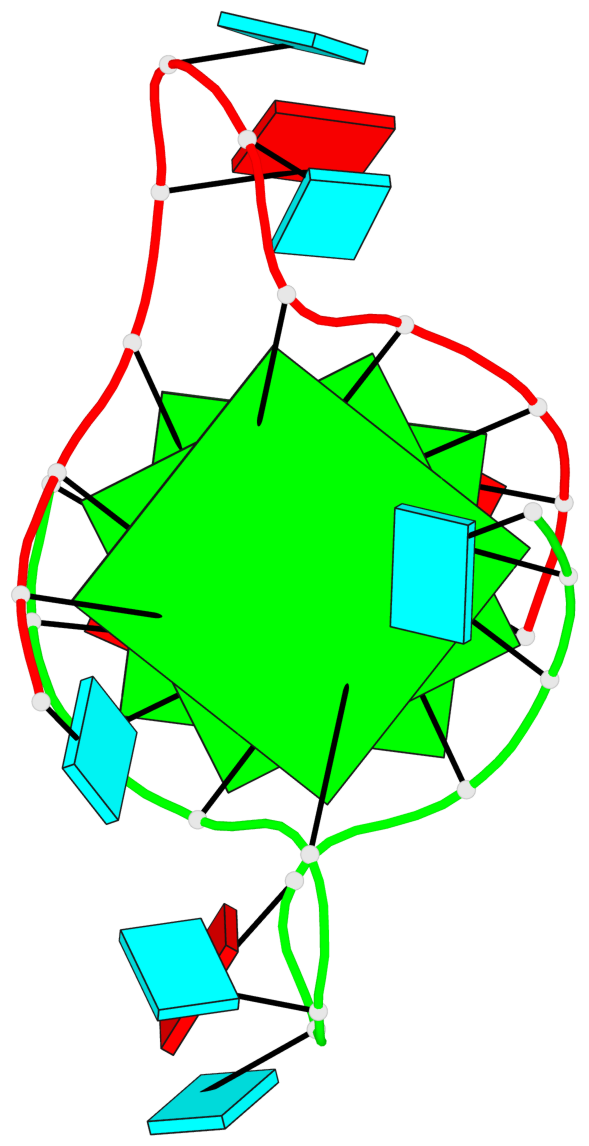
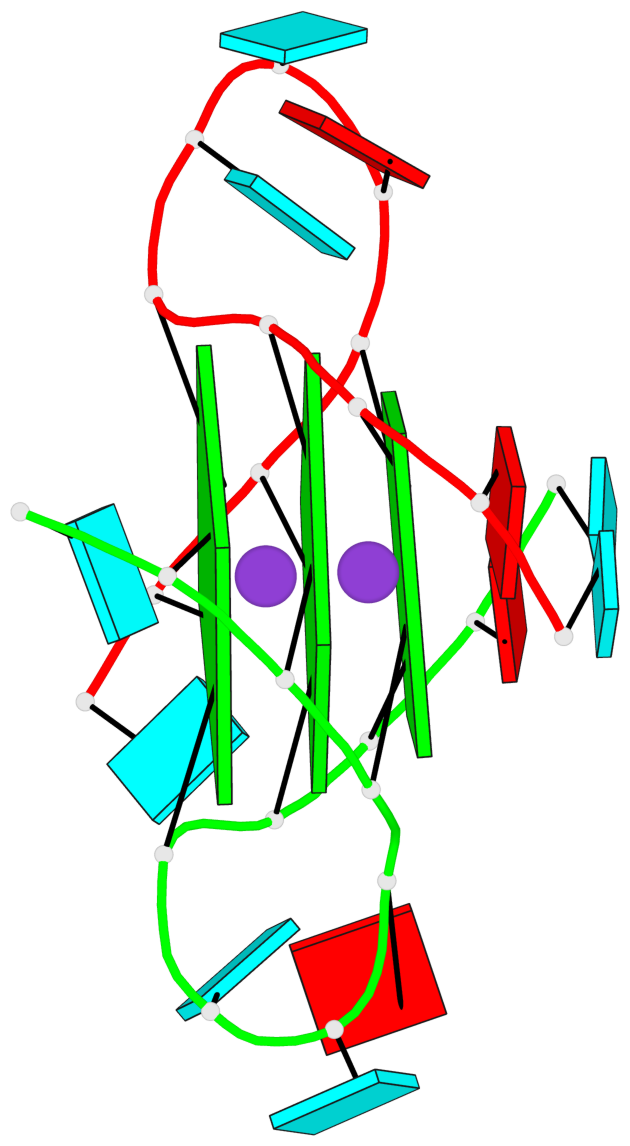
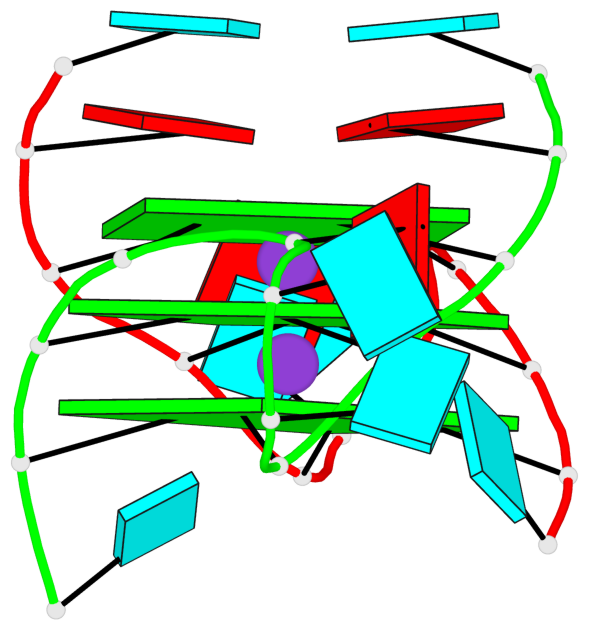
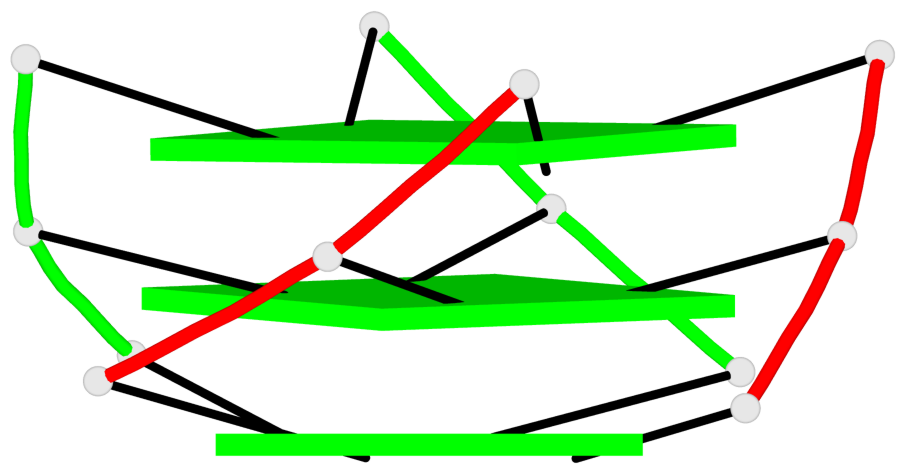
Base-block schematics in six views
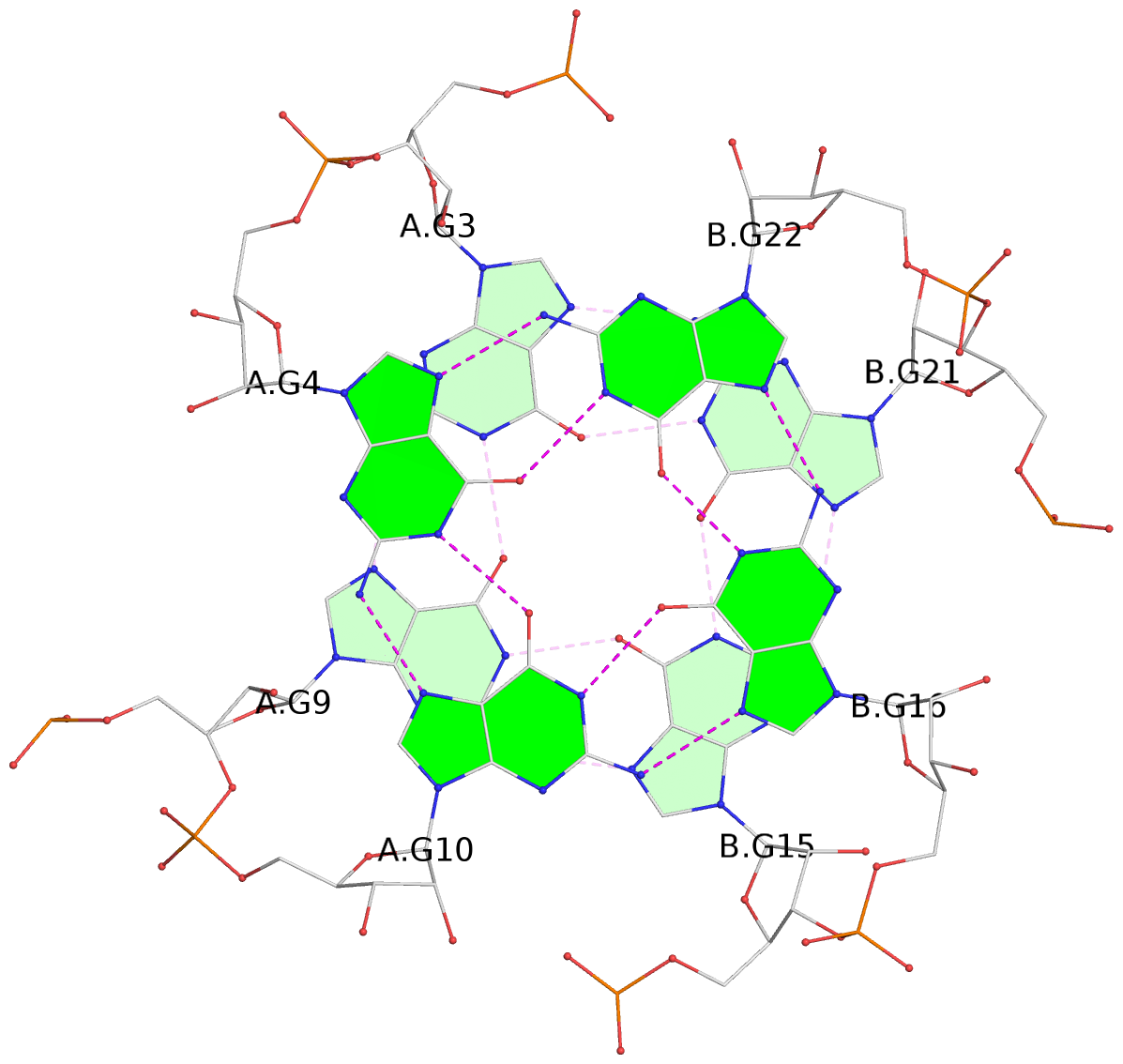
List of 3 G-tetrads
1 glyco-bond=---- sugar=--33 groove=---- planarity=0.167 type=other nts=4 GGGG A.G3,A.G9,B.G15,B.G21 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.100 type=planar nts=4 GGGG A.G4,A.G10,B.G16,B.G22 3 glyco-bond=---- sugar=---3 groove=---- planarity=0.288 type=bowl nts=4 GGGG A.G5,A.G11,B.G17,B.G23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.