Detailed DSSR results for the G-quadruplex: PDB entry 3qsc
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 3qsc
- Class
- DNA
- Method
- X-ray (2.4 Å)
- Summary
- The first crystal structure of a human telomeric g-quadruplex DNA bound to a metal-containing ligand (a copper complex)
- Reference
- Campbell NH, Karim NH, Parkinson GN, Gunaratnam M, Petrucci V, Todd AK, Vilar R, Neidle S (2012): "Molecular basis of structure-activity relationships between salphen metal complexes and human telomeric DNA quadruplexes." J.Med.Chem., 55, 209-222. doi: 10.1021/jm201140v.
- Abstract
- The first X-ray crystal structures of nickel(II) and copper(II) salphen metal complexes bound to a quadruplex DNA are presented. Two structures have been determined and show that these salphen-metal complexes bind to human telomeric quadruplexes by end-stacking, with the metal in each case almost in line with the potassium ion channel. Quadruplex and duplex DNA binding is presented for these two and other related salphen complexes, all with side-chains terminating in pyrrolidino end-groups and differing patterns of substitution on the salphen core. The crystal structures are able to provide rationalizations for the structure-activity data, and in particular for the superior quadruplex-binding of the nickel complexes compared to that of the copper-containing ones. The complexes show significant antiproliferative activity for the compounds in a panel of cancer cell lines. They also show telomerase inhibitory activity in the telomerase TRAP-LIG assay.
- G4 notes
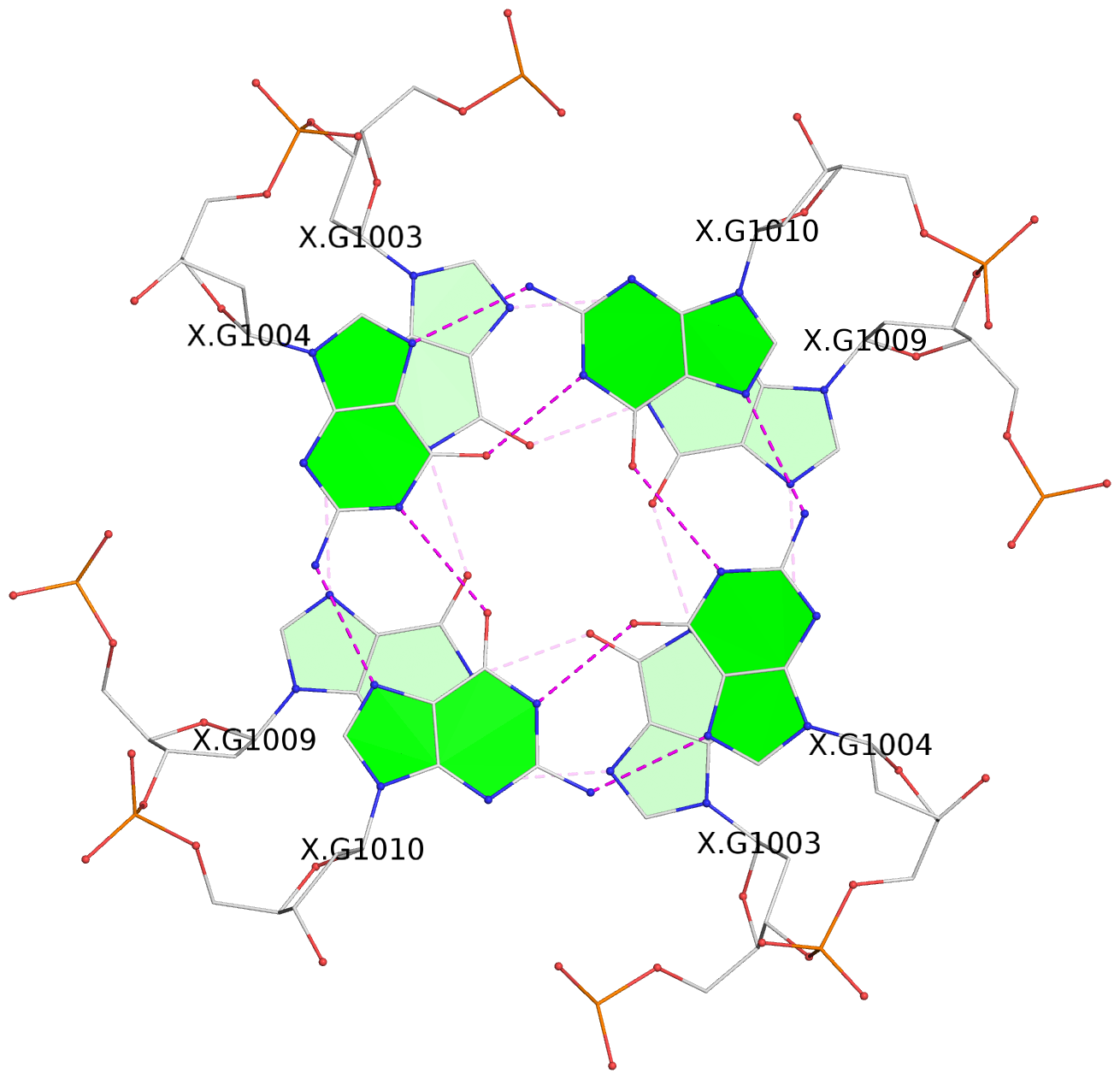
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
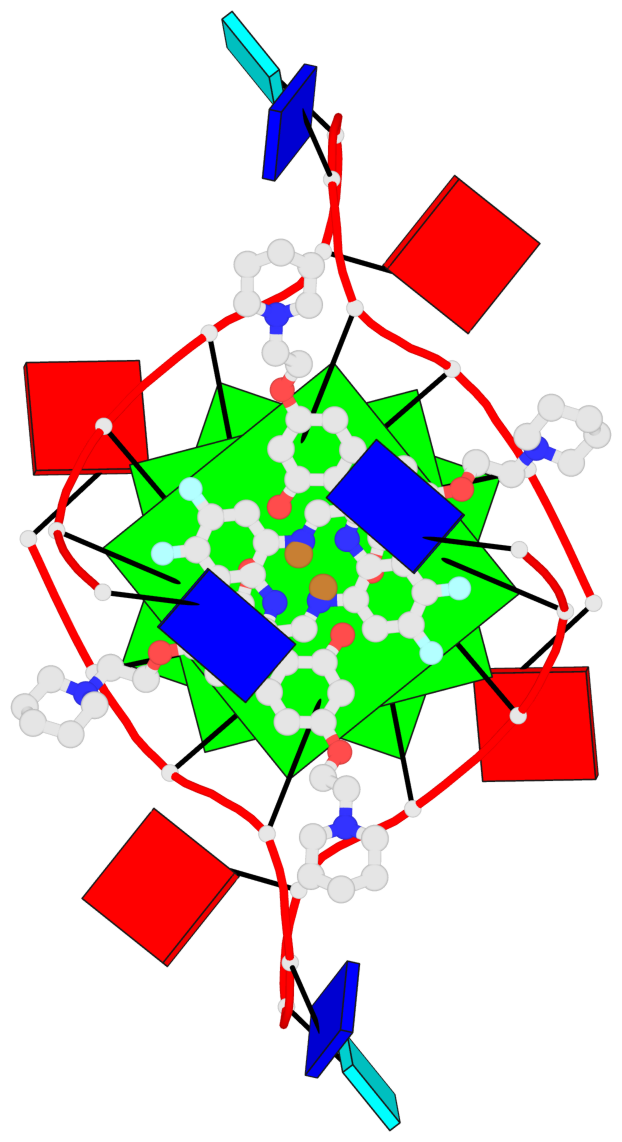
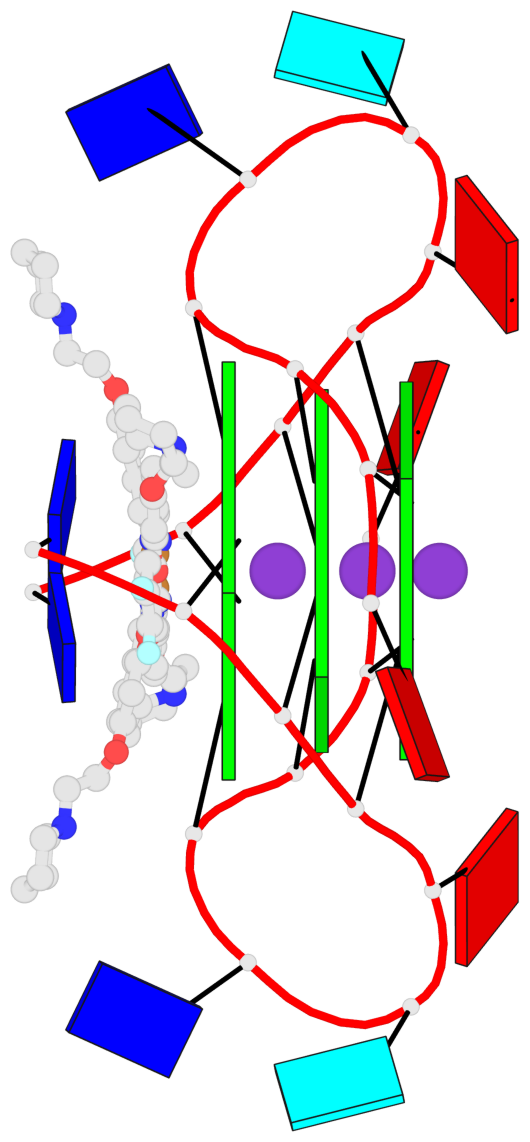
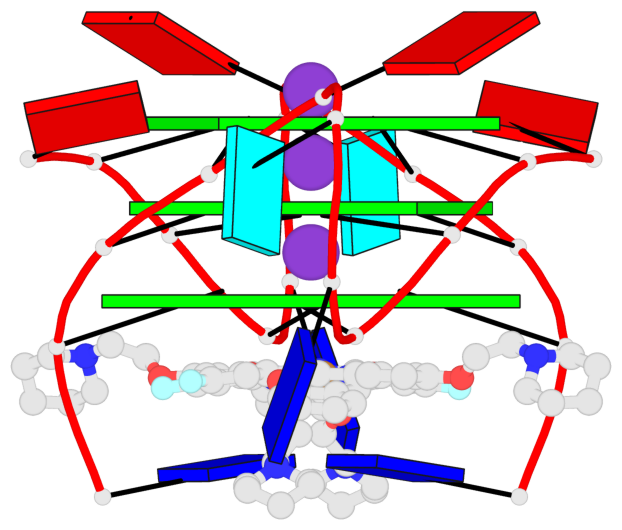
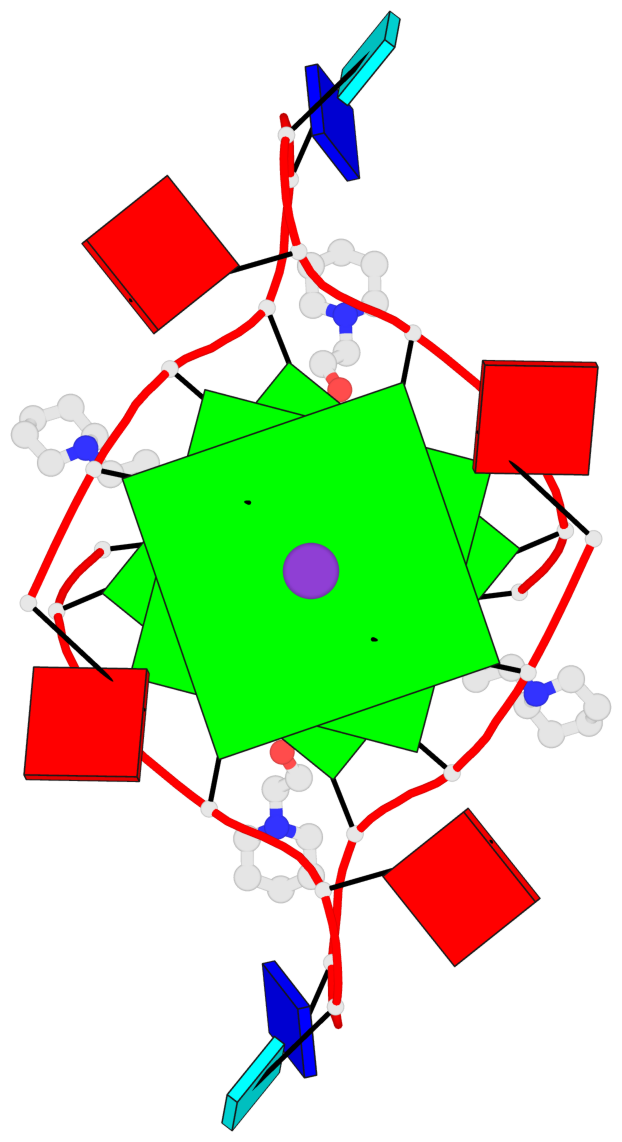
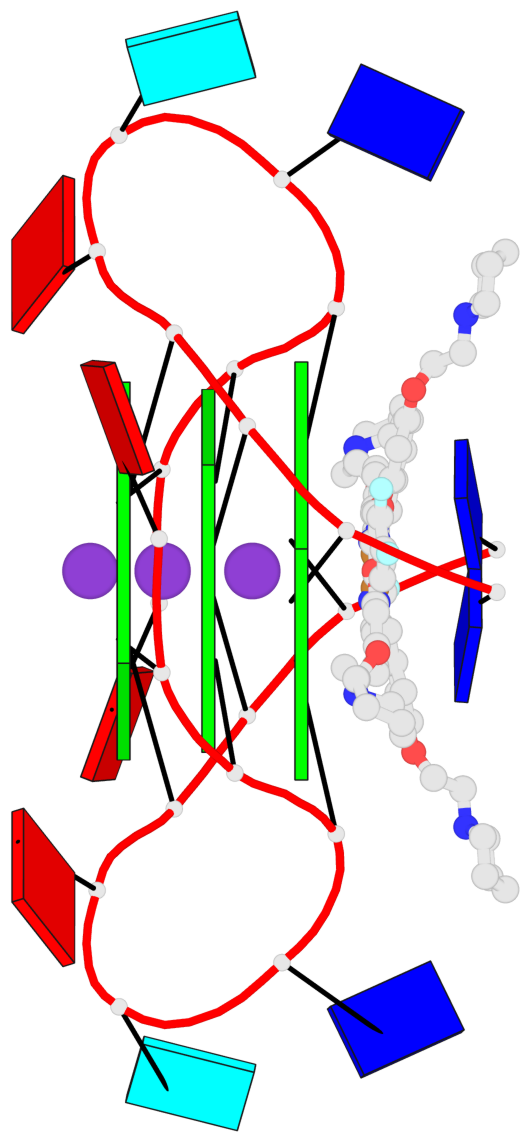
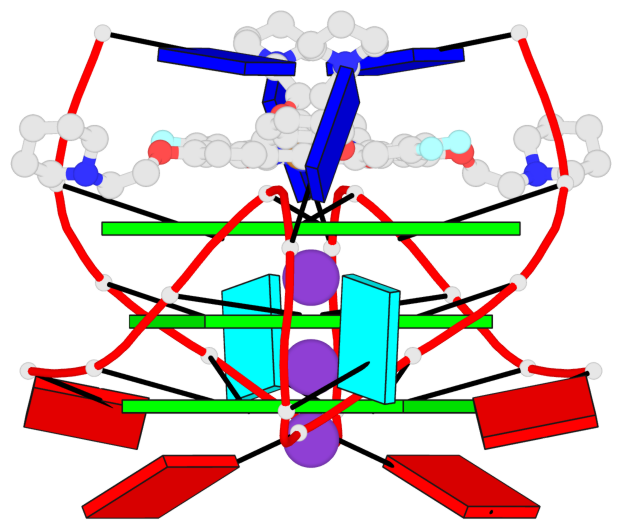
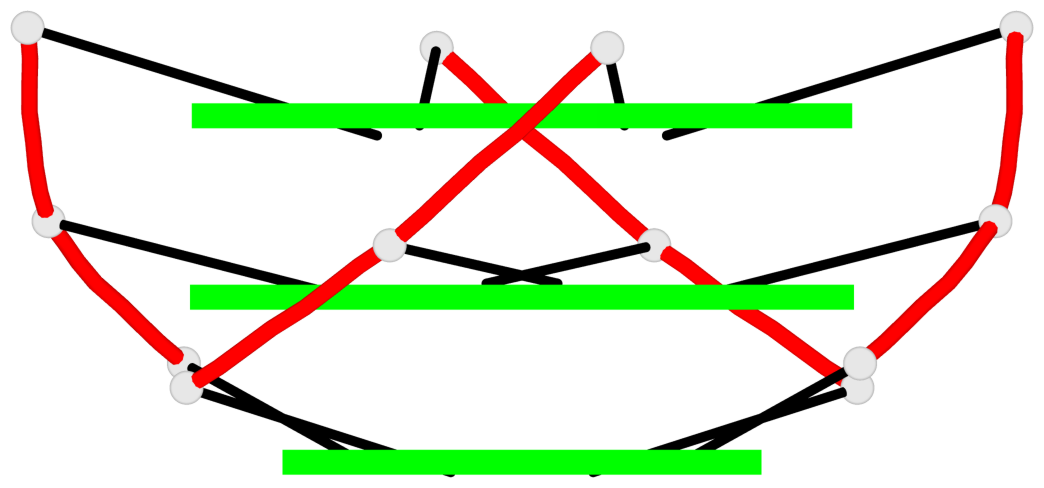
Base-block schematics in six views
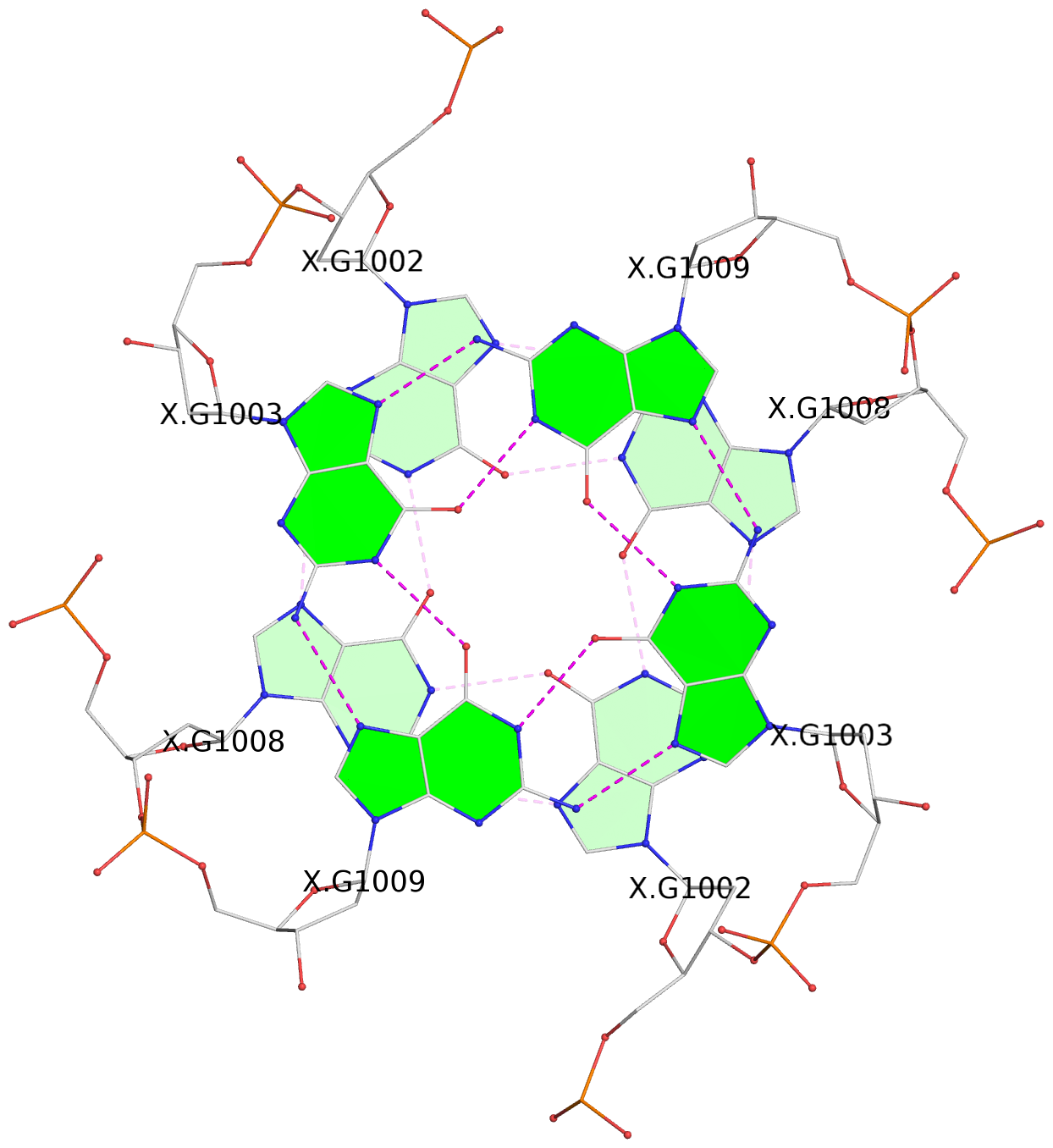
List of 3 G-tetrads
1 glyco-bond=---- sugar=3-3- groove=---- planarity=0.430 type=bowl-2 nts=4 GGGG 1:X.DG1002,1:X.DG1008,2:X.DG1002,2:X.DG1008 2 glyco-bond=---- sugar=3.3. groove=---- planarity=0.298 type=saddle nts=4 GGGG 1:X.DG1003,1:X.DG1009,2:X.DG1003,2:X.DG1009 3 glyco-bond=---- sugar=---- groove=---- planarity=0.302 type=bowl nts=4 GGGG 1:X.DG1004,1:X.DG1010,2:X.DG1004,2:X.DG1010
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.