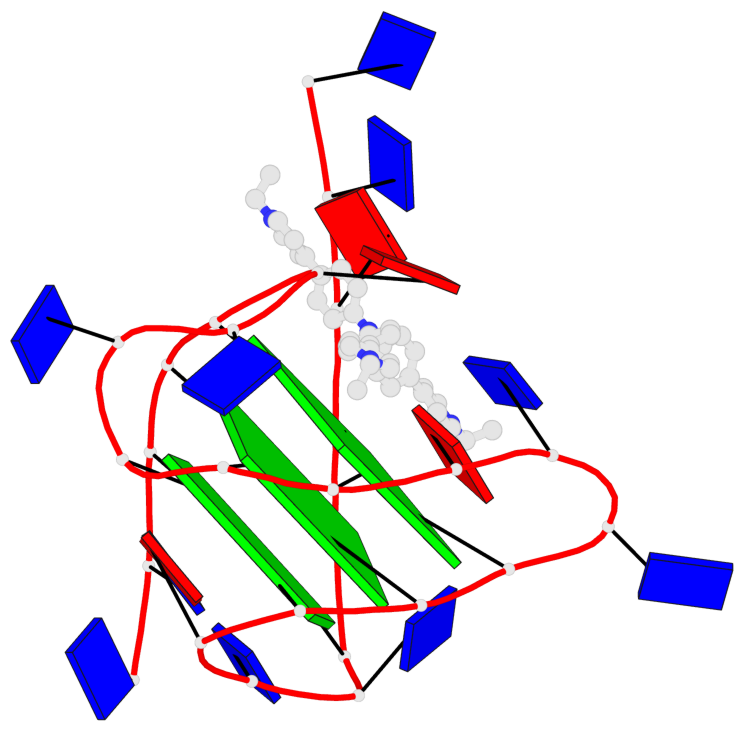
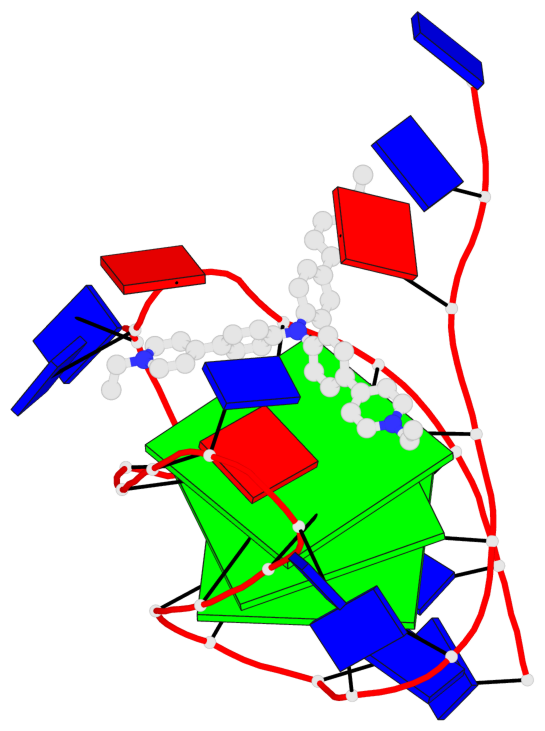
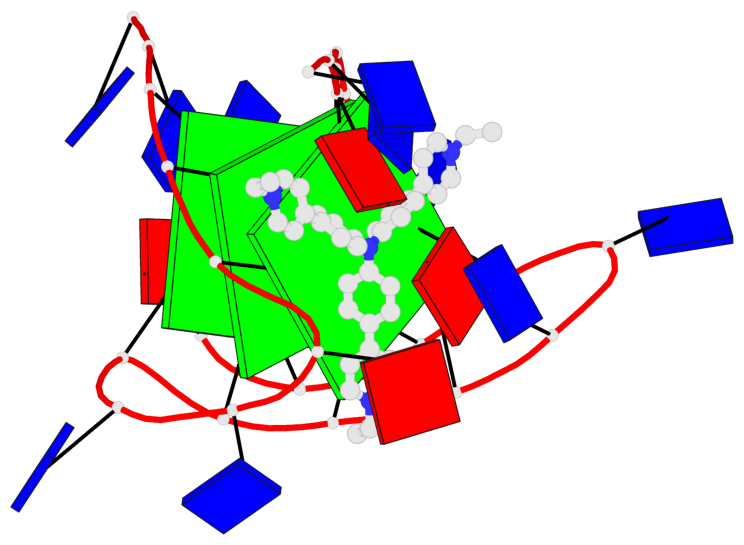
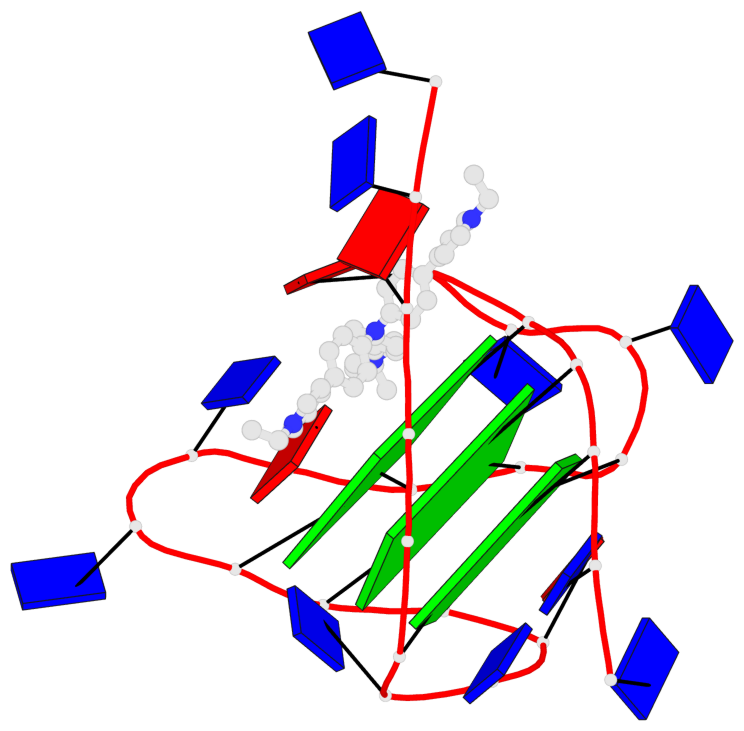
Detailed DSSR results for the G-quadruplex: PDB entry 6kfj
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6kfj
- Class
- DNA
- Method
- NMR
- Summary
- NMR solution structure of the 1:1 complex of wttel26 g-quadruplex and a tripodal cationic fluorescent probe nbte
- Reference
- Liu LY, Liu W, Wang KN, Zhu BC, Xia XY, Ji LN, Mao ZW (2020): "Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination." Angew.Chem.Int.Ed.Engl., 59, 9719-9726. doi: 10.1002/anie.202002422.
- Abstract
- G-quadruplex DNA show structural polymorphism, leading to challenges in the use of selective recognition probes for the accurate detection of G-quadruplexes in vivo. Herein, we present a tripodal cationic fluorescent probe, NBTE, which showed distinguishable fluorescence lifetime responses between G-quadruplexes and other DNA topologies, and fluorescence quantum yield (Φf ) enhancement upon G-quadruplex binding. We determined two NBTE-G-quadruplex complex structures with high Φf values by NMR spectroscopy. The structures indicated NBTE interacted with G-quadruplexes using three arms through π-π stacking, differing from that with duplex DNA using two arms, which rationalized the higher Φf values and lifetime response of NBTE upon G-quadruplex binding. Based on photon counts of FLIM, we detected the percentage of G-quadruplex DNA in live cells with NBTE and found G-quadruplex DNA content in cancer cells is 4-fold that in normal cells, suggesting the potential applications of this probe in cancer cell detection.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
Base-block schematics in six views
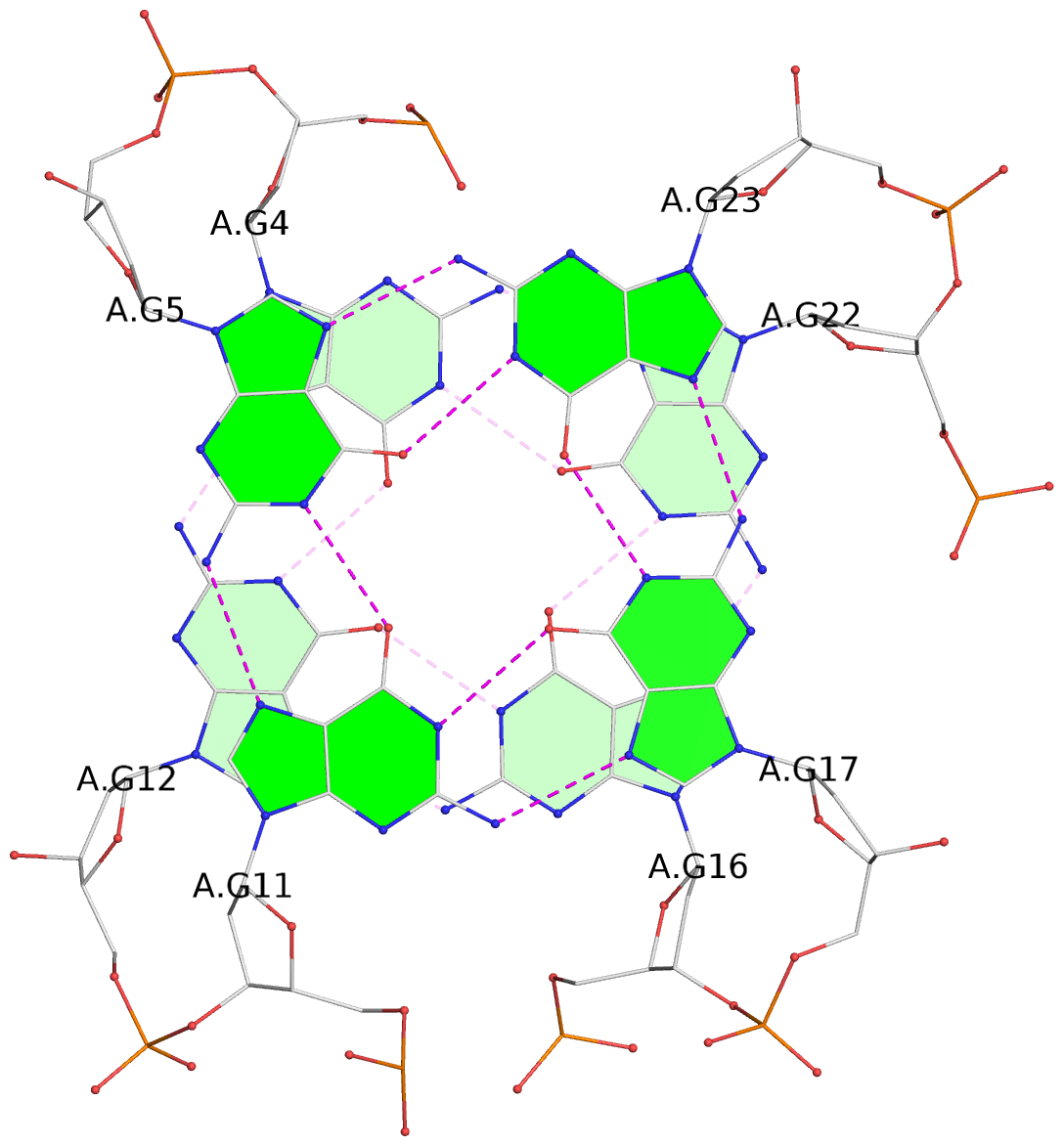
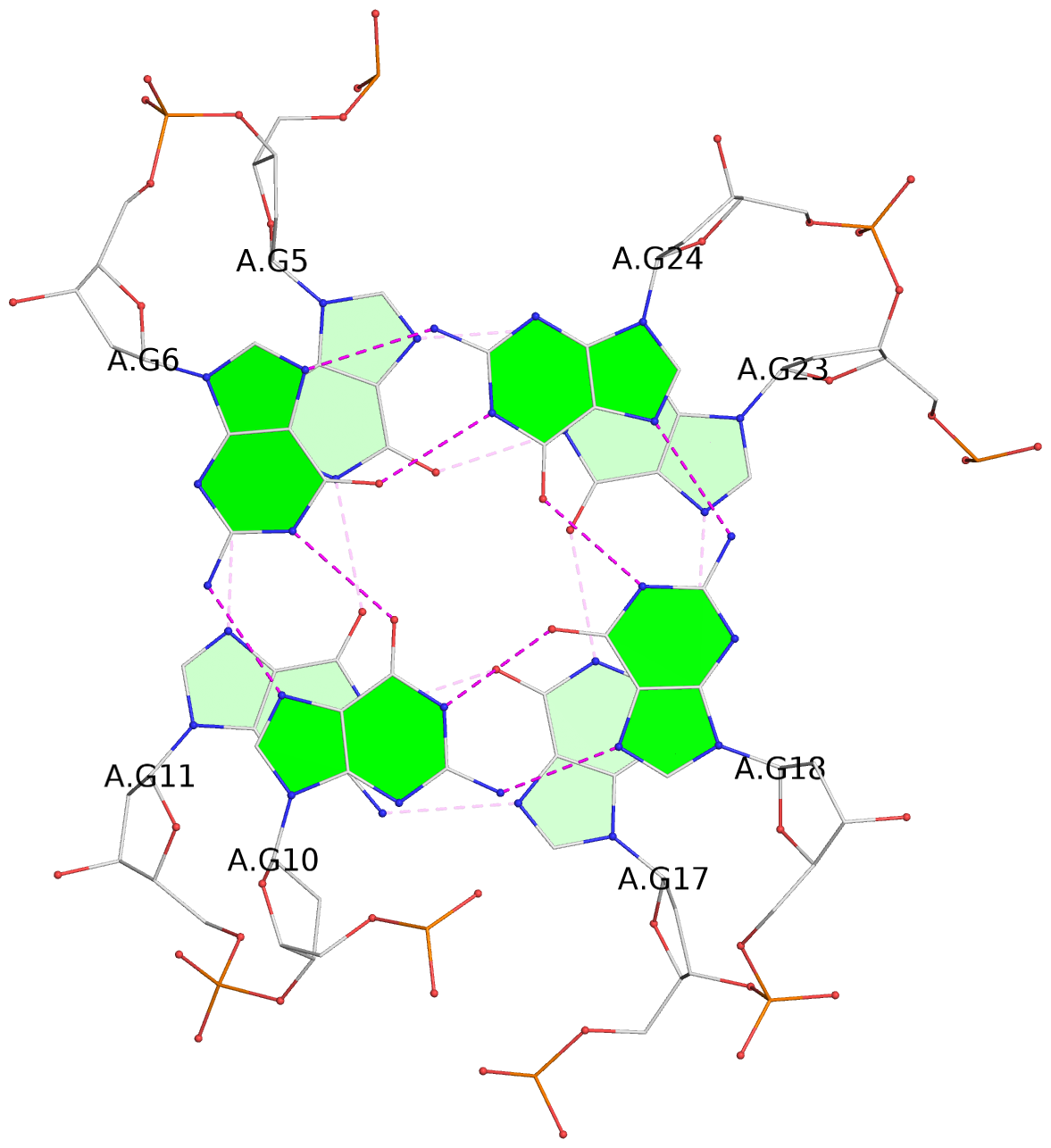
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=-.-- groove=wn-- planarity=0.218 type=other nts=4 GGGG A.DG4,A.DG12,A.DG16,A.DG22 2 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.208 type=other nts=4 GGGG A.DG5,A.DG11,A.DG17,A.DG23 3 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.165 type=other nts=4 GGGG A.DG6,A.DG10,A.DG18,A.DG24
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.