Detailed DSSR results for the G-quadruplex: PDB entry 6m05
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6m05
- Class
- DNA
- Method
- NMR
- Summary
- Trimolecular g-quadruplex
- Reference
- Jing H, Fu W, Hu W, Xu S, Xu X, He M, Liu Y, Zhang N (2021): "NMR structural study on the self-trimerization of d(GTTAGG) into a dynamic trimolecular G-quadruplex assembly preferentially in Na+ solution with a moderate K+ tolerance." Nucleic Acids Res., 49, 2306-2316. doi: 10.1093/nar/gkab028.
- Abstract
- Vast G-quadruplexes (GQs) are primarily folded by one, two, or four G-rich oligomers, rarely with an exception. Here, we present the first NMR solution structure of a trimolecular GQ (tri-GQ) that is solely assembled by the self-trimerization of d(GTTAGG), preferentially in Na+ solution tolerant to an equal amount of K+ cation. Eight guanines from three asymmetrically folded strands of d(GTTAGG) are organized into a two-tetrad core, which features a broken G-column and two width-irregular grooves. Fast strand exchanges on a timescale of second at 17°C spontaneously occur between folded tri-GQ and unfolded single-strand of d(GTTAGG) that both species coexist in dynamic equilibrium. Thus, this tri-GQ is not just simply a static assembly but rather a dynamic assembly. Moreover, another minor tetra-GQ that has putatively tetrameric (2+2) antiparallel topology becomes noticeable only at an extremely high strand concentration above 18 mM. The major tri-GQ and minor tetra-GQ are considered to be mutually related, and their reversible interconversion pathways are proposed accordingly. The sequence d(GTTAGG) could be regarded as either a reading frame shifted single repeat of human telomeric DNA or a 1.5 repeat of Bombyx mori telomeric DNA. Overall, our findings provide new insight into GQs and expect more functional applications.
- G4 notes
- 2 G-tetrads, 1 G4 helix
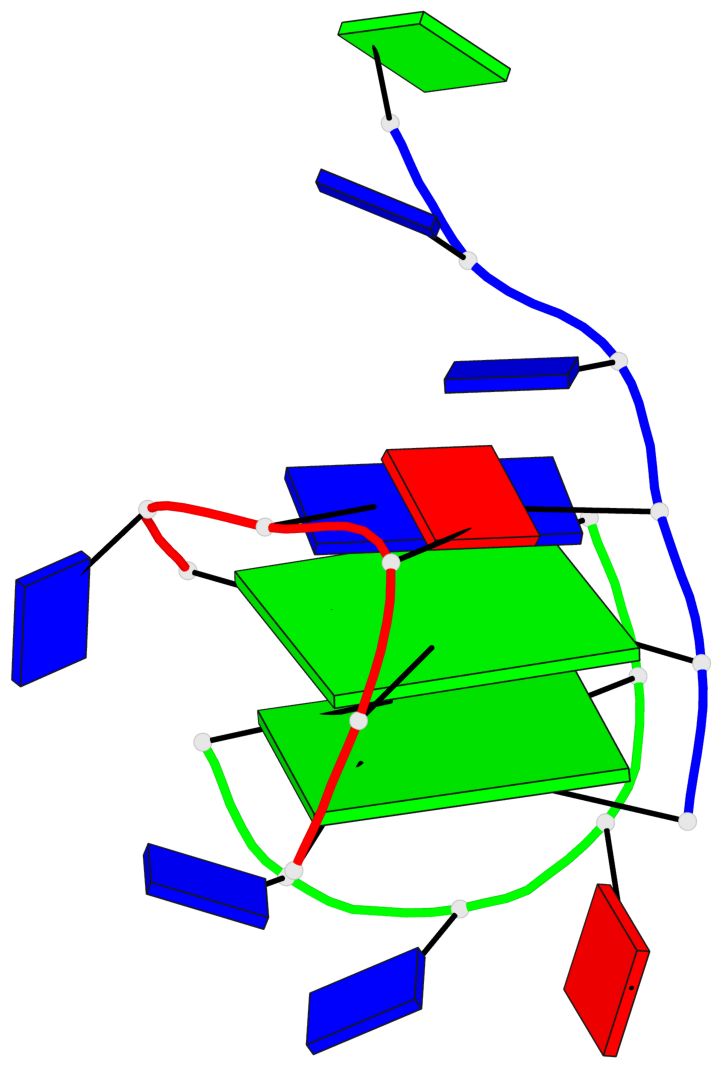
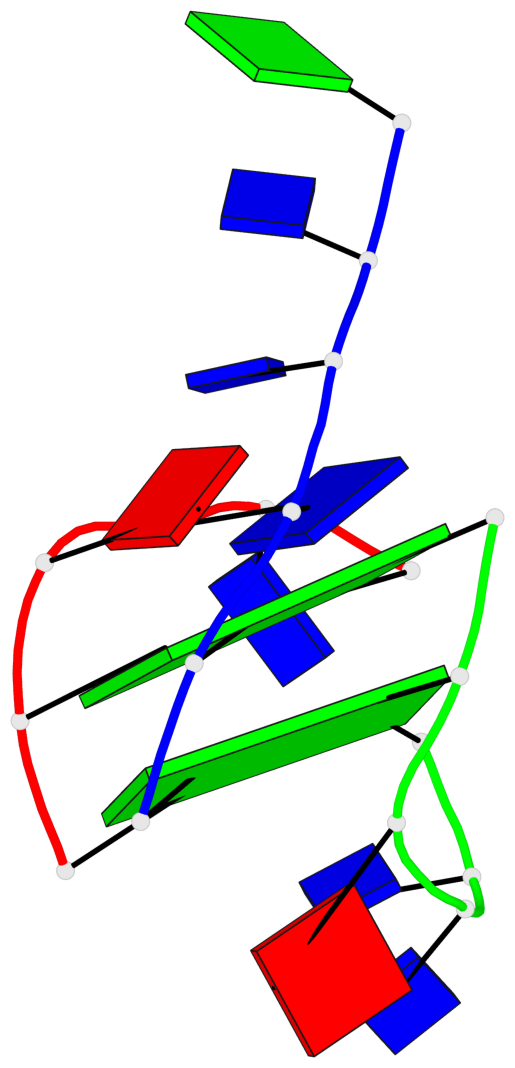
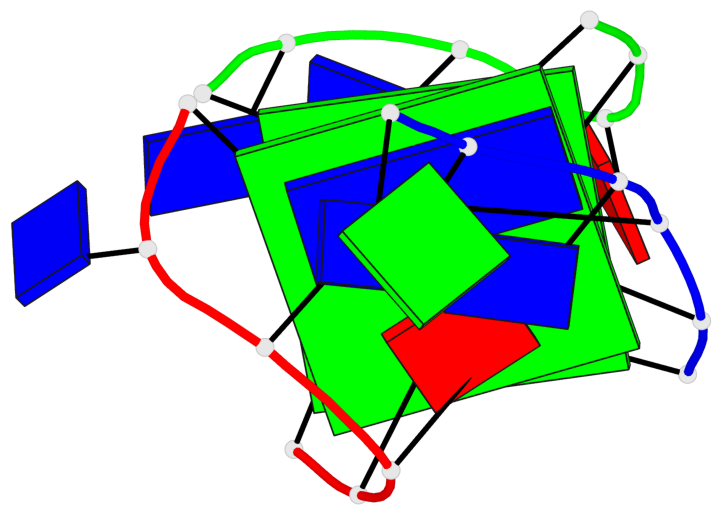
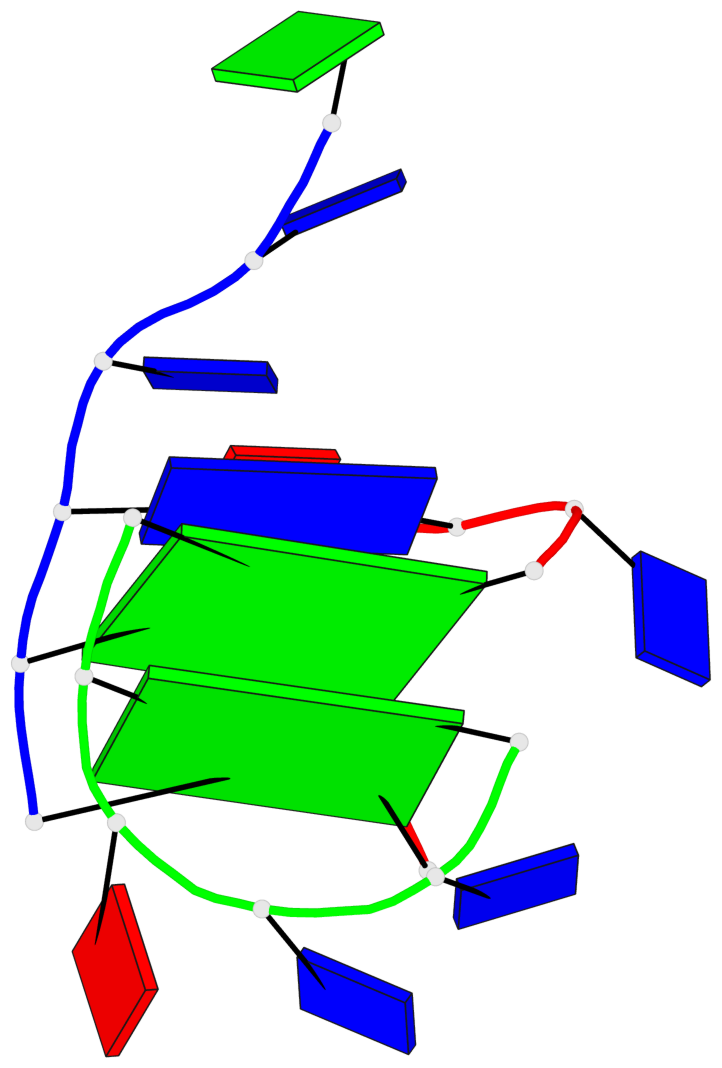
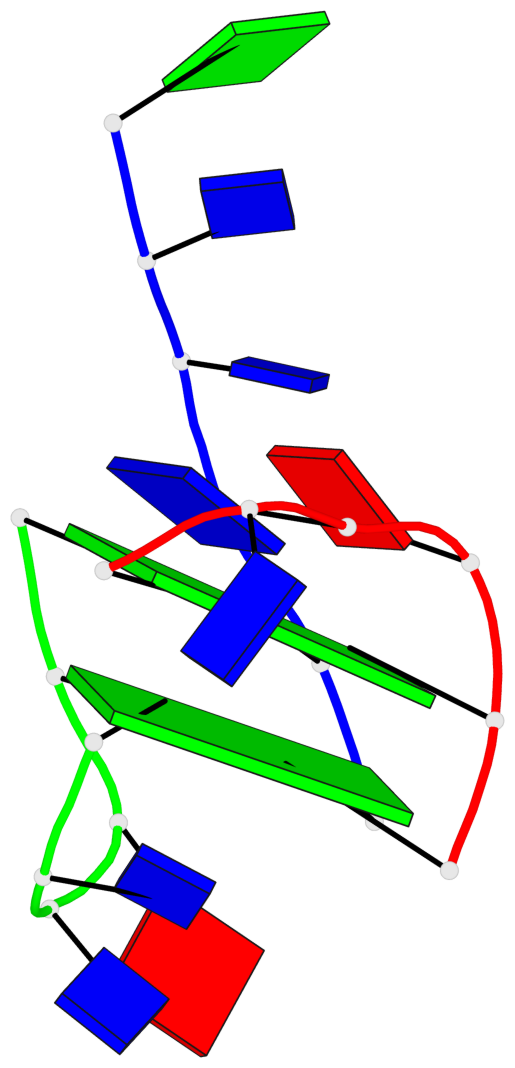
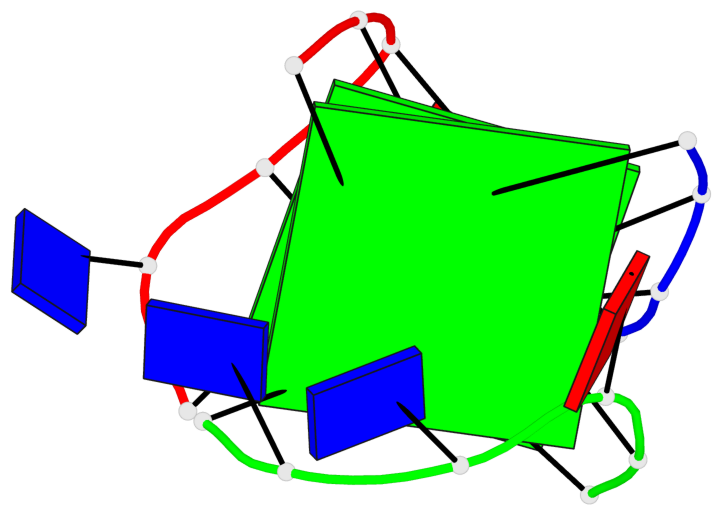
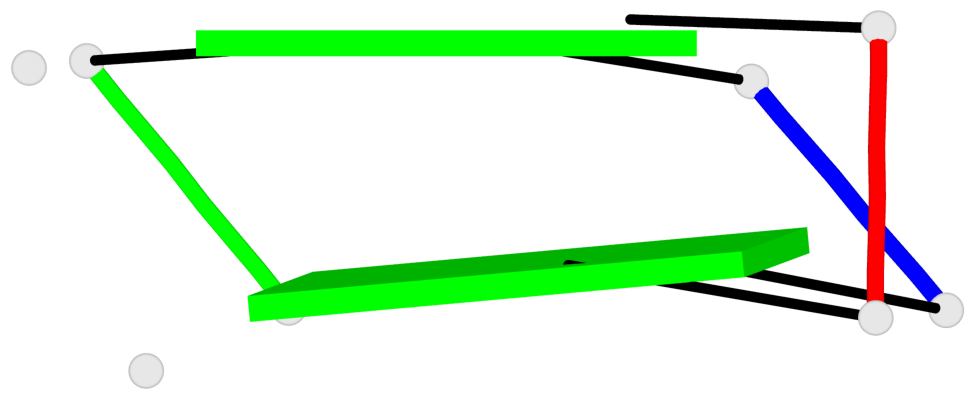
Base-block schematics in six views
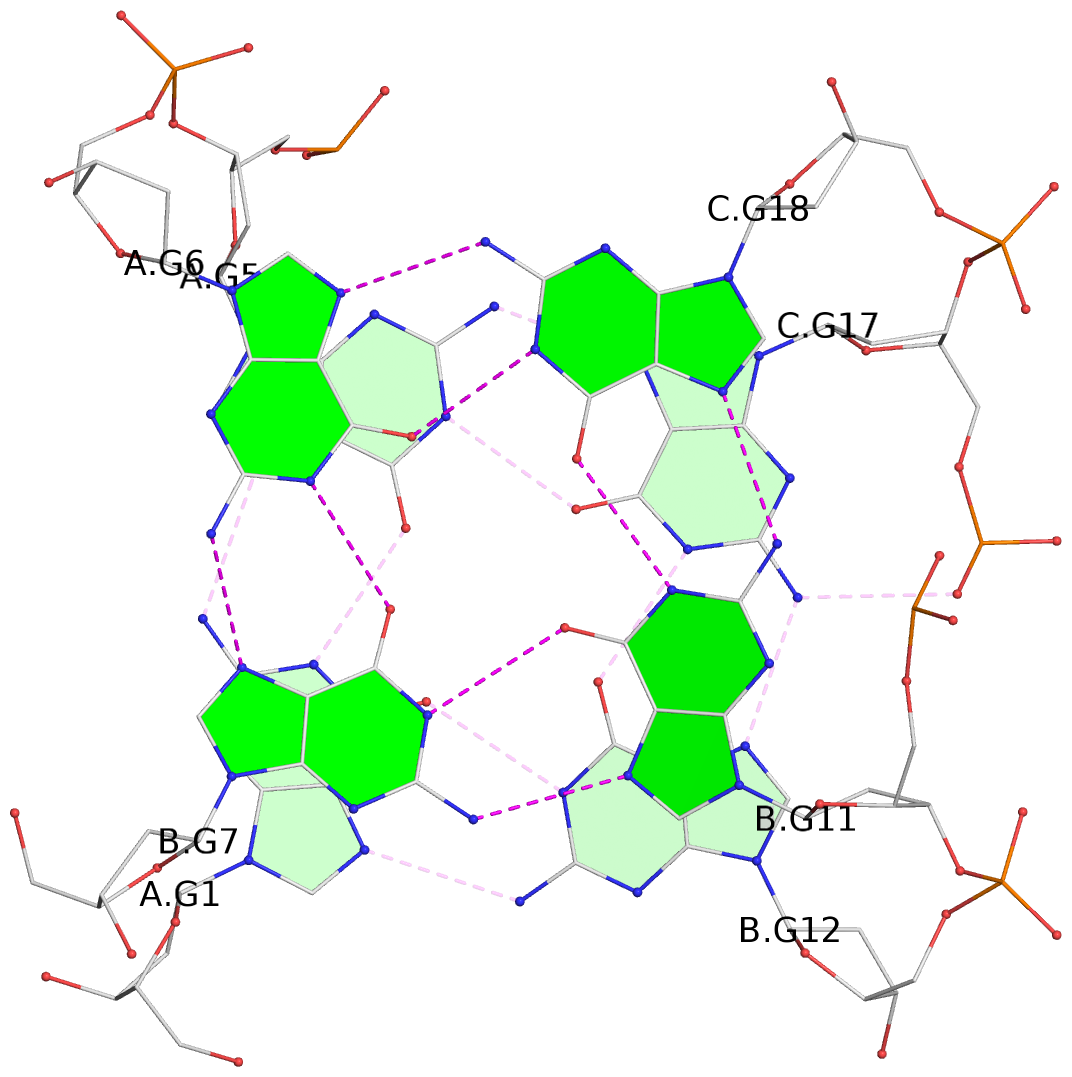
List of 2 G-tetrads
1 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.236 type=other nts=4 GGGG A.DG1,A.DG5,C.DG17,B.DG12 2 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.448 type=bowl nts=4 GGGG A.DG6,B.DG7,B.DG11,C.DG18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, inter-molecular
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=5 GTTAG A.DG1,A.DT2,A.DT3,A.DA4,A.DG5 2 type=lateral helix=#1 nts=5 GTTAG B.DG7,B.DT8,B.DT9,B.DA10,B.DG11