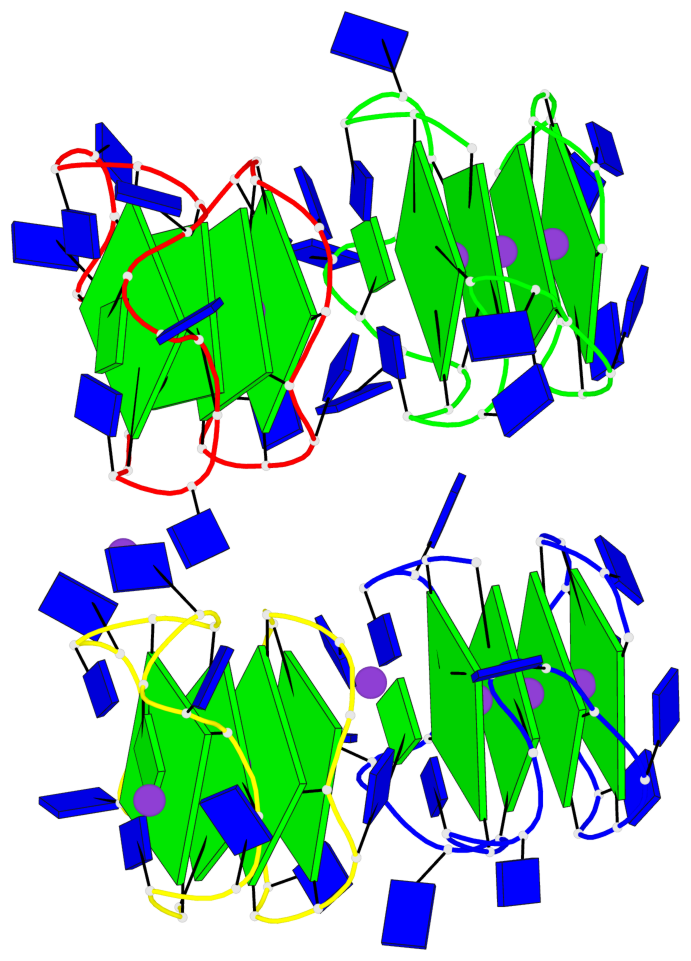
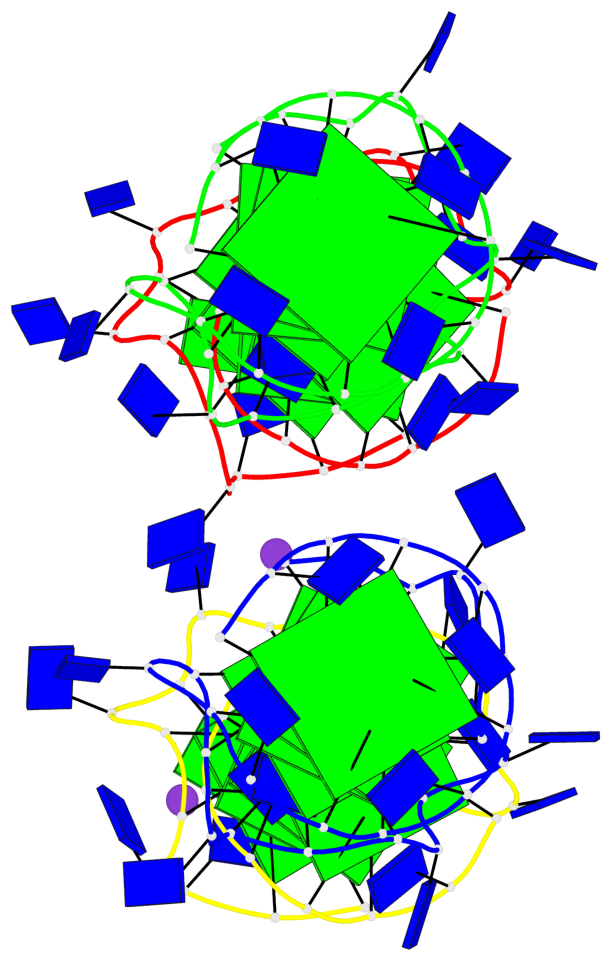
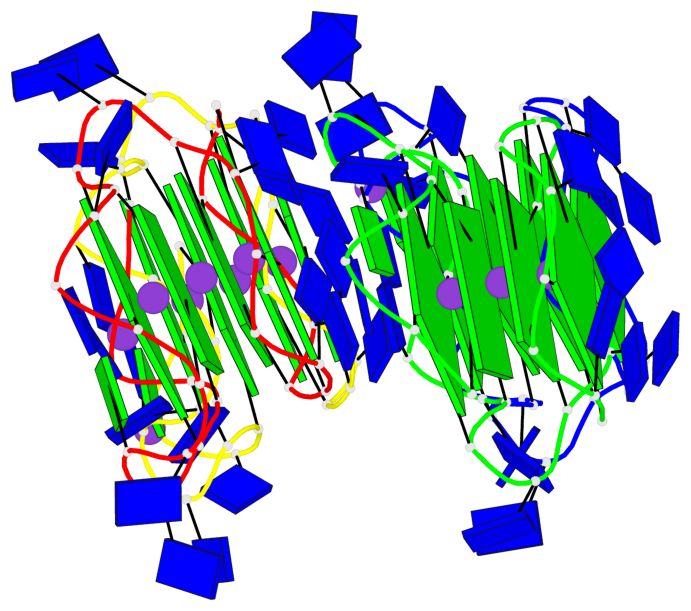
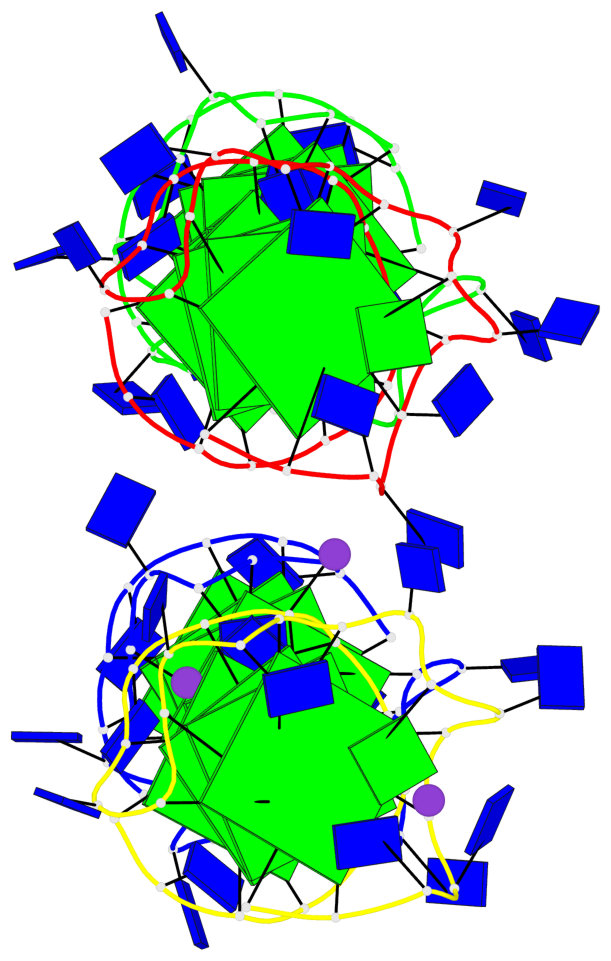
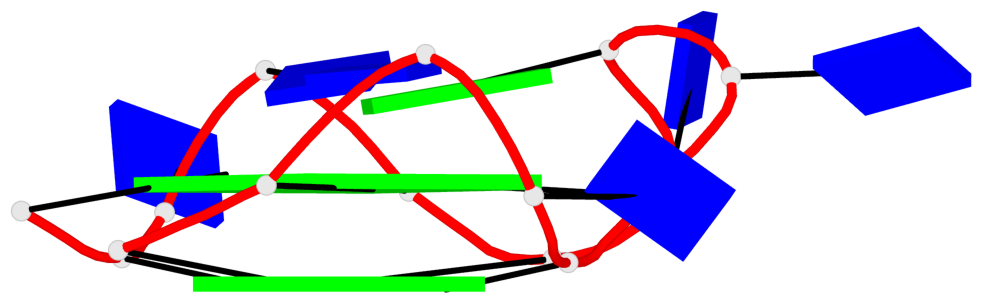
Detailed DSSR results for the G-quadruplex: PDB entry 6qjo
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6qjo
- Class
- DNA
- Method
- X-ray (1.8 Å)
- Summary
- DNA containing both right- and left-handed parallel-stranded g-quadruplexes
- Reference
- Winnerdy FR, Bakalar B, Maity A, Vandana JJ, Mechulam Y, Schmitt E, Phan AT (2019): "NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes." Nucleic Acids Res., 47, 8272-8281. doi: 10.1093/nar/gkz349.
- Abstract
- Analogous to the B- and Z-DNA structures in double-helix DNA, there exist both right- and left-handed quadruple-helix (G-quadruplex) DNA. Numerous conformations of right-handed and a few left-handed G-quadruplexes were previously observed, yet they were always identified separately. Here, we present the NMR solution and X-ray crystal structures of a right- and left-handed hybrid G-quadruplex. The structure reveals a stacking interaction between two G-quadruplex blocks with different helical orientations and displays features of both right- and left-handed G-quadruplexes. An analysis of loop mutations suggests that single-nucleotide loops are preferred or even required for the left-handed G-quadruplex formation. The discovery of a right- and left-handed hybrid G-quadruplex further expands the polymorphism of G-quadruplexes and is potentially useful in designing a left-to-right junction in G-quadruplex engineering.
- G4 notes
- 16 G-tetrads, 4 G4 helices, 4 G4 stems, 2(-P-P-P), parallel(4+0), UUUU
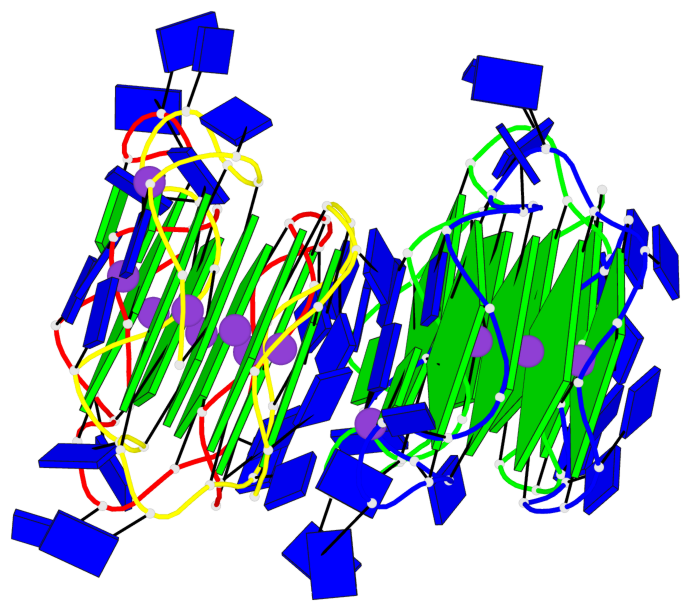
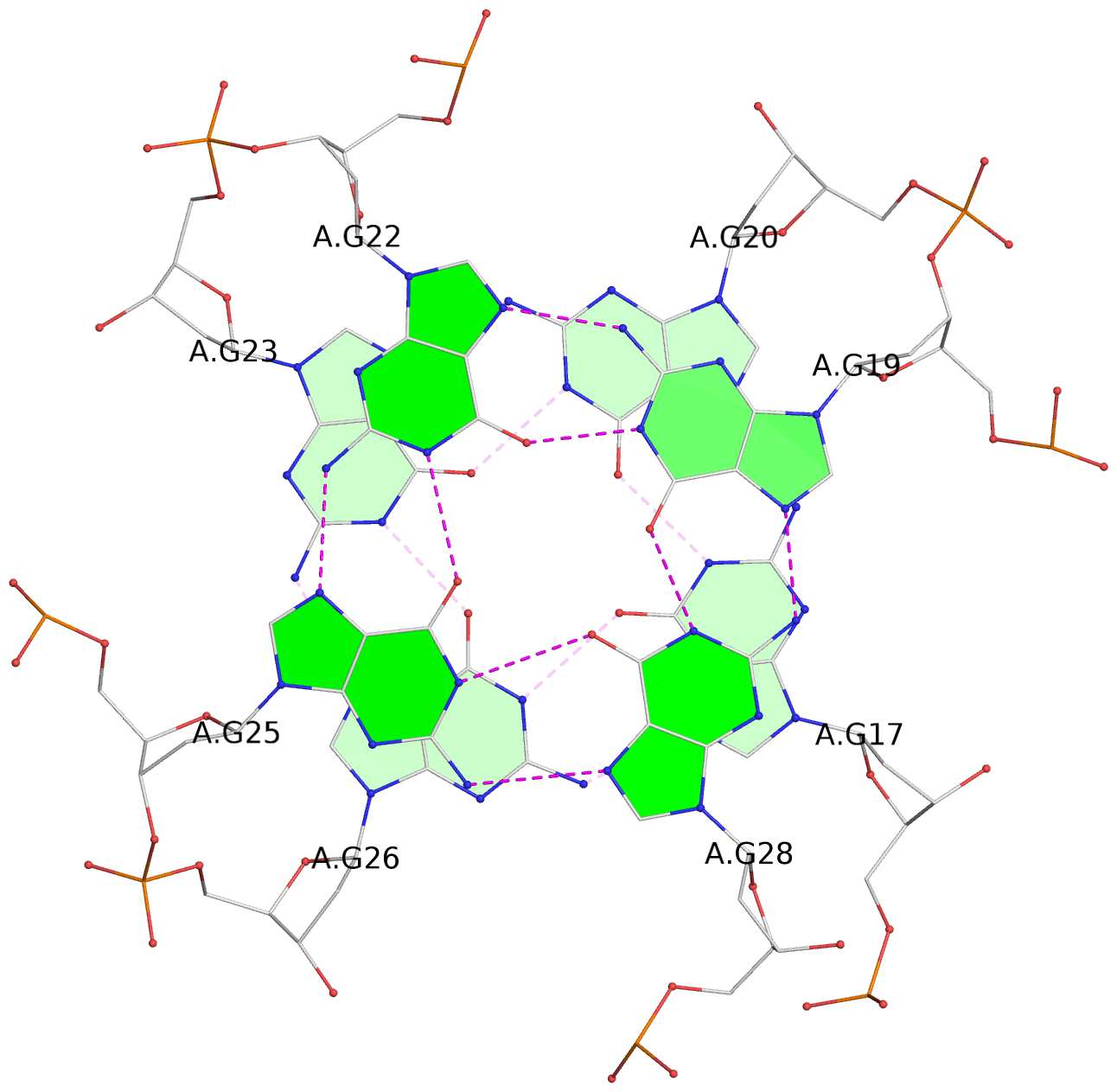
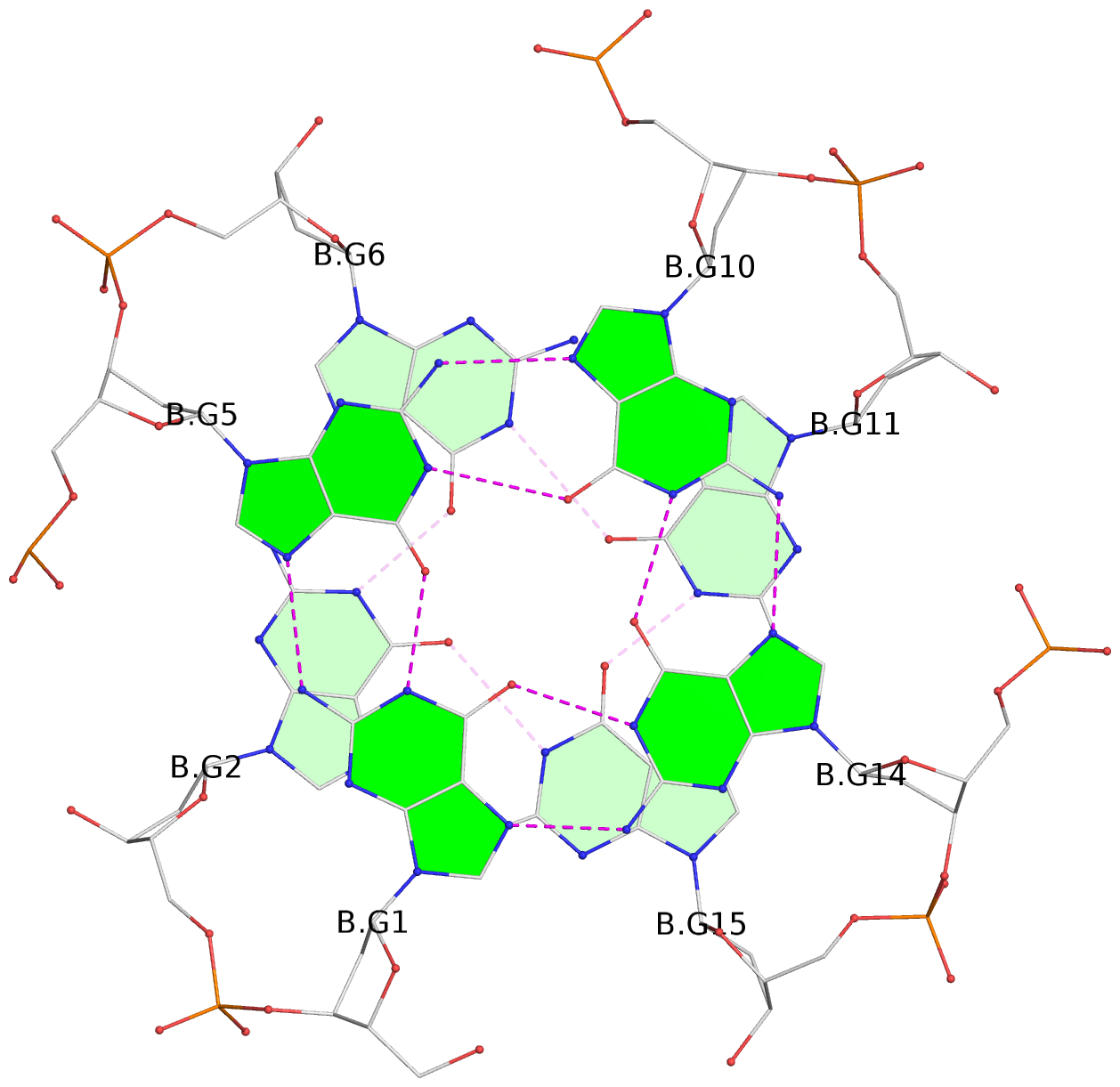
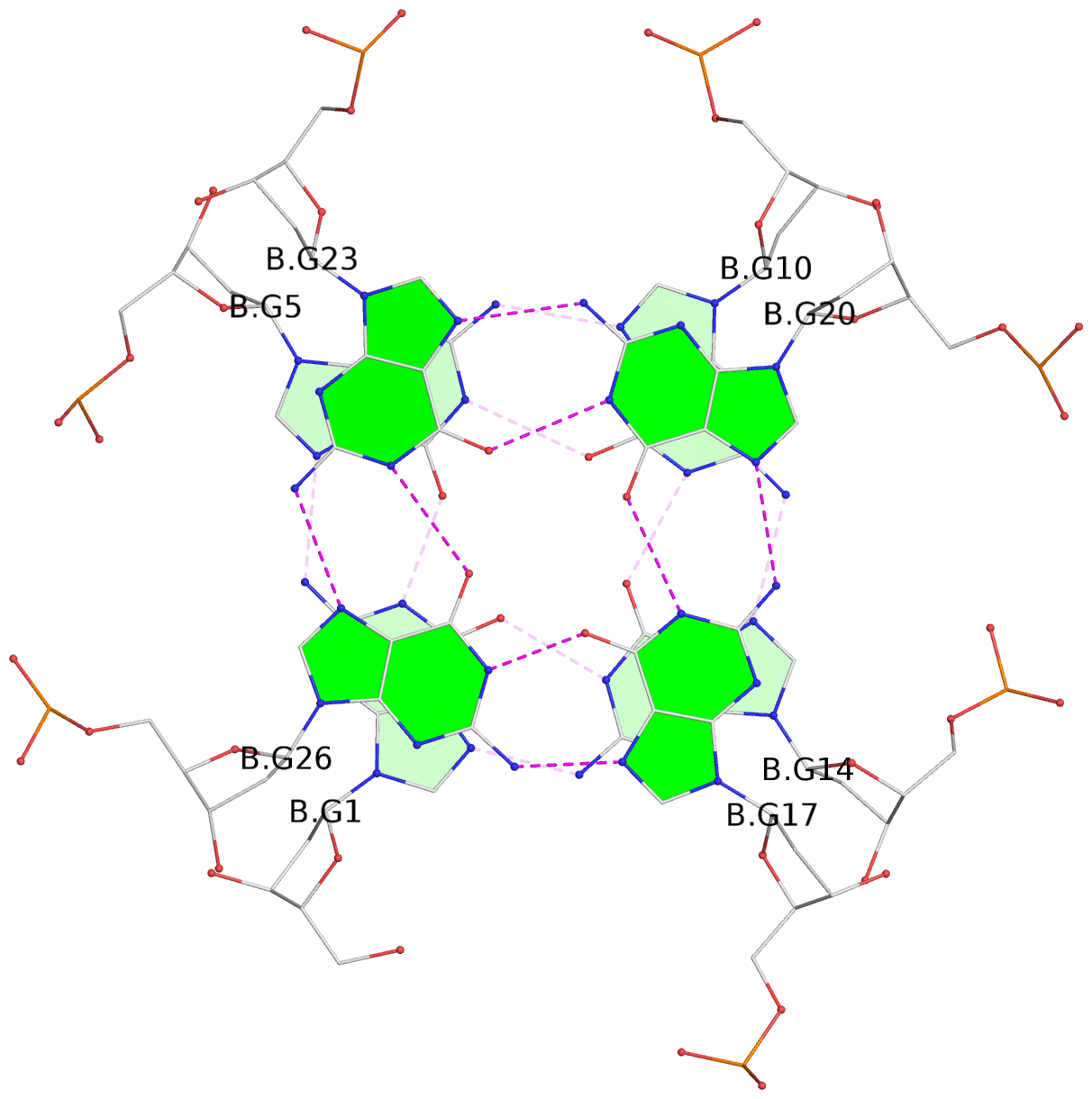
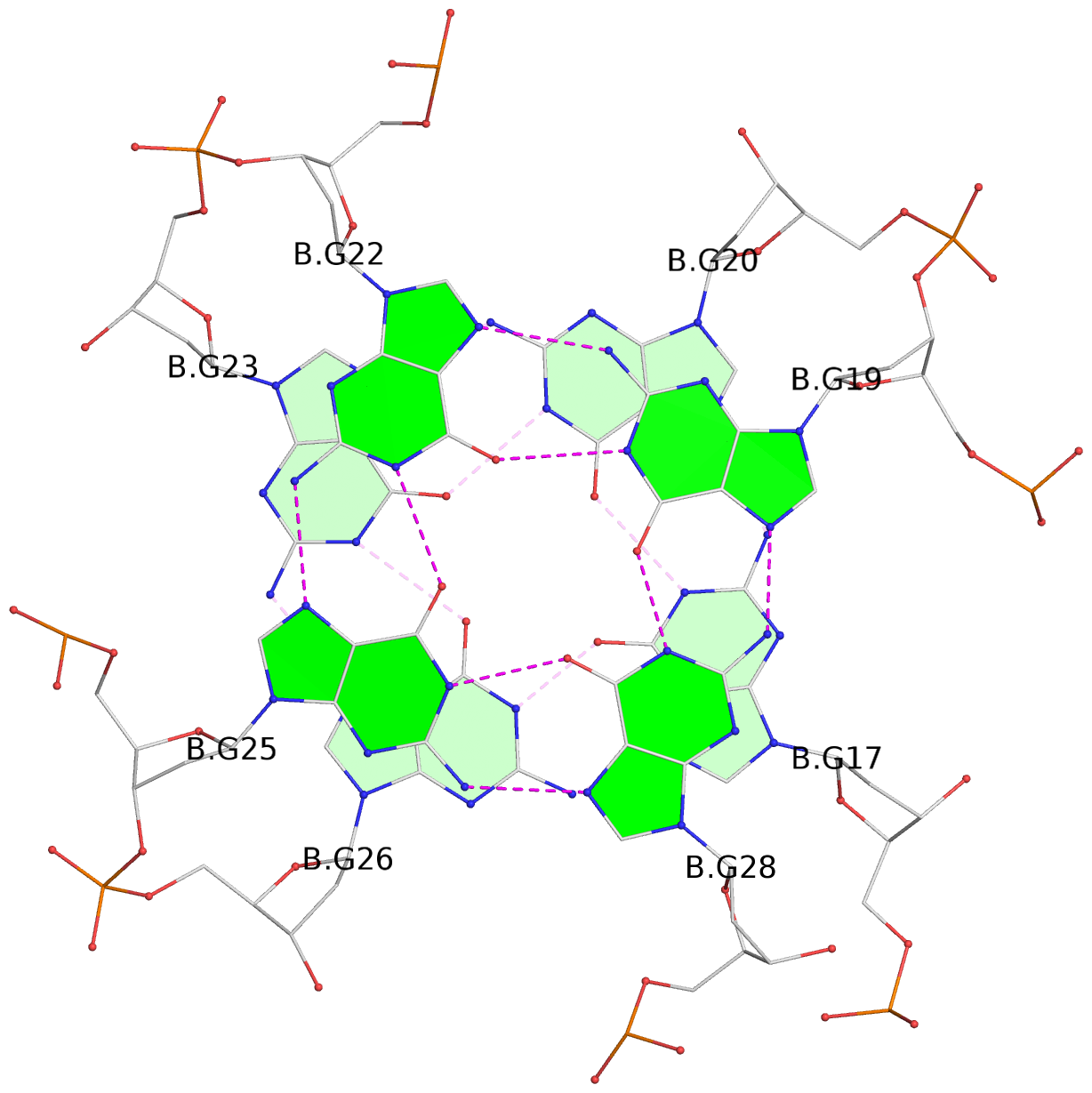
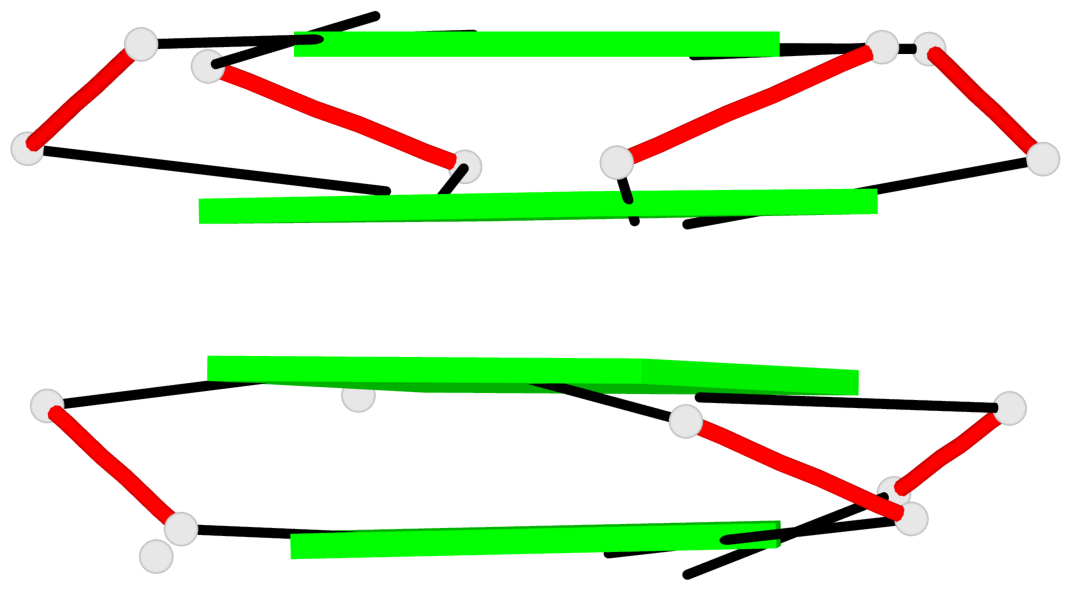
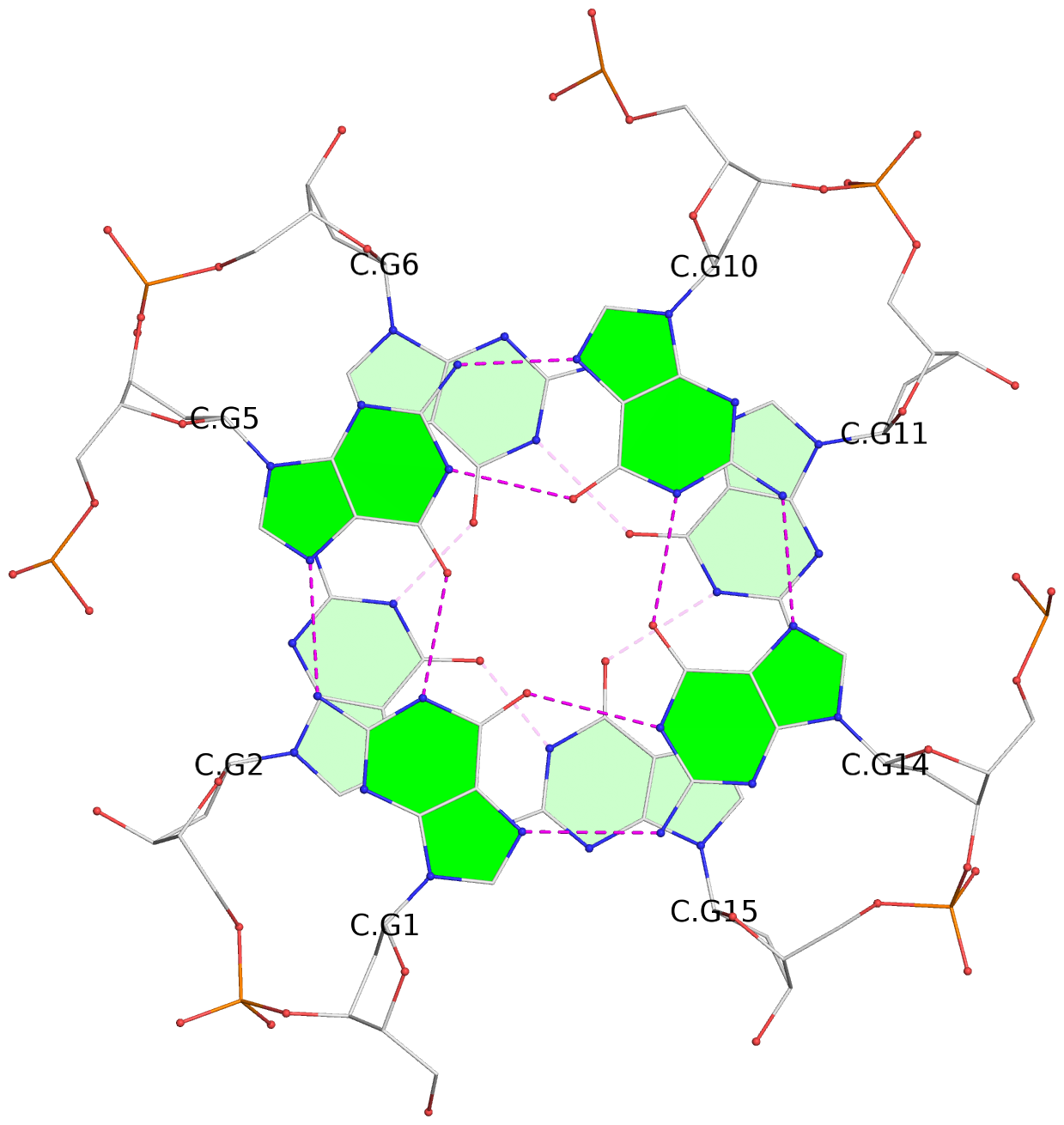
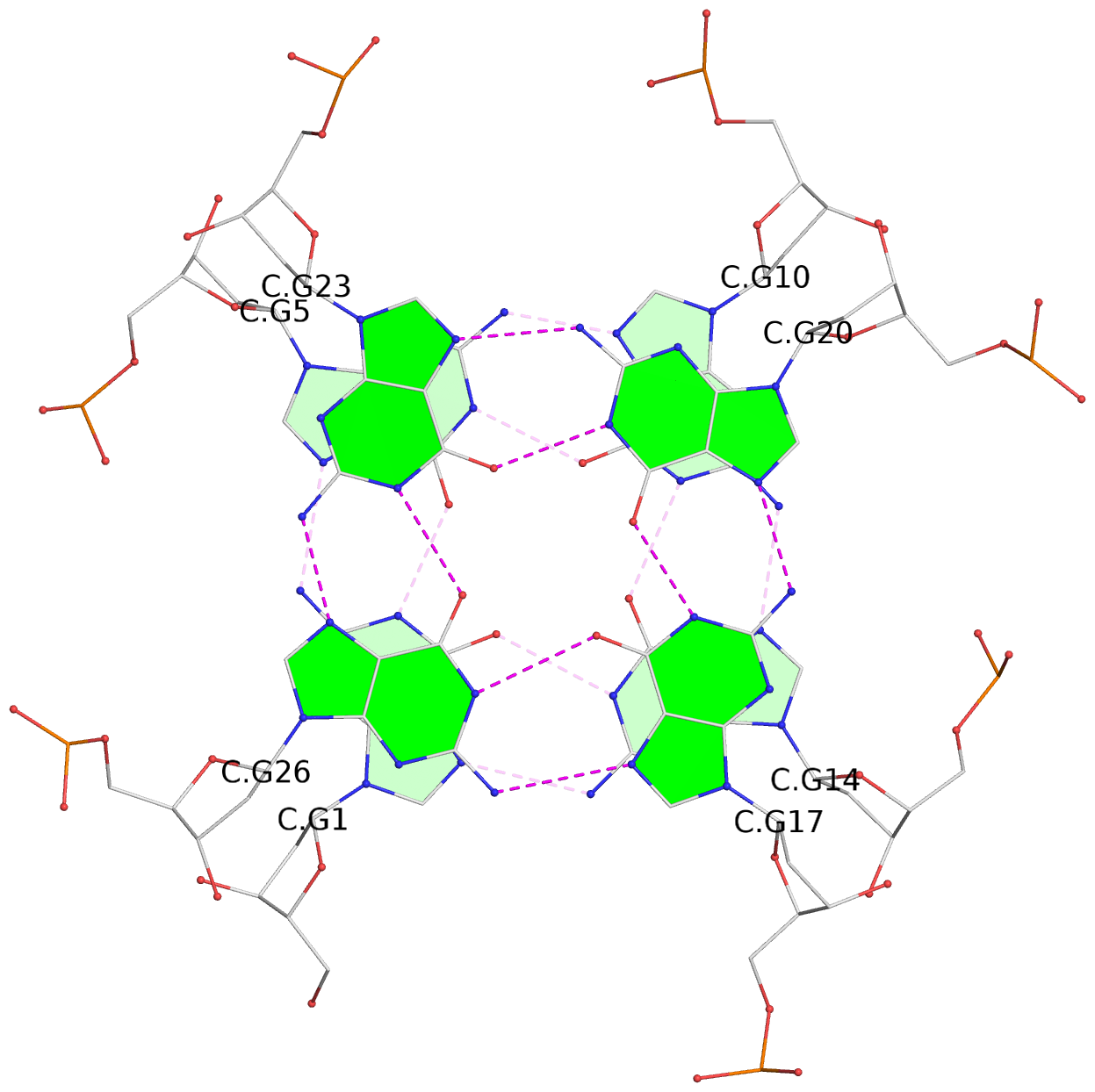
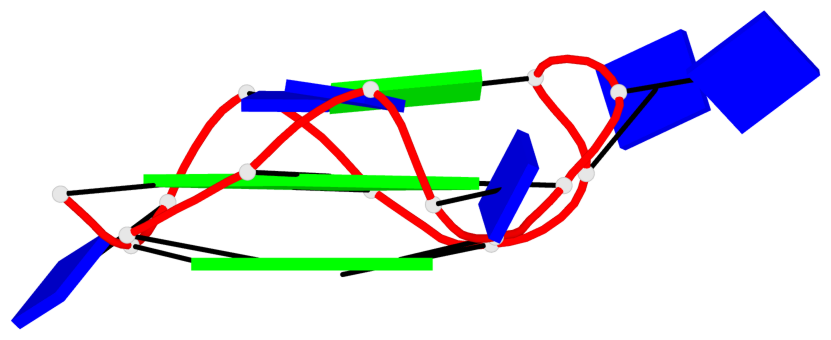
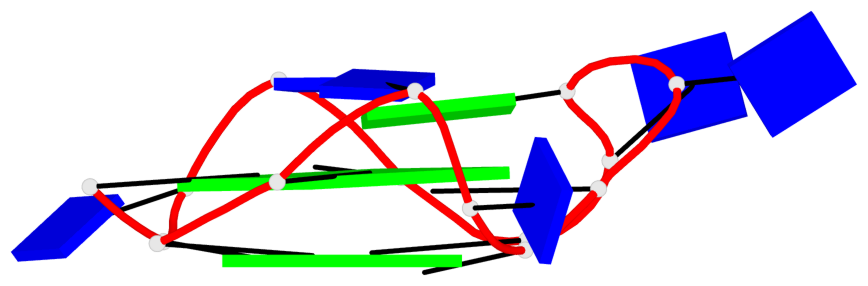
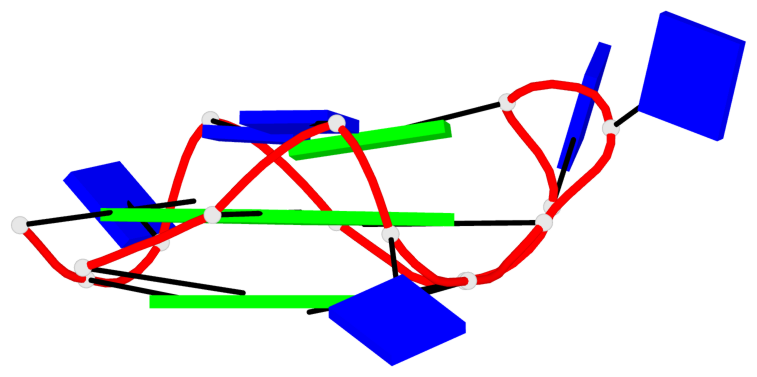
Base-block schematics in six views
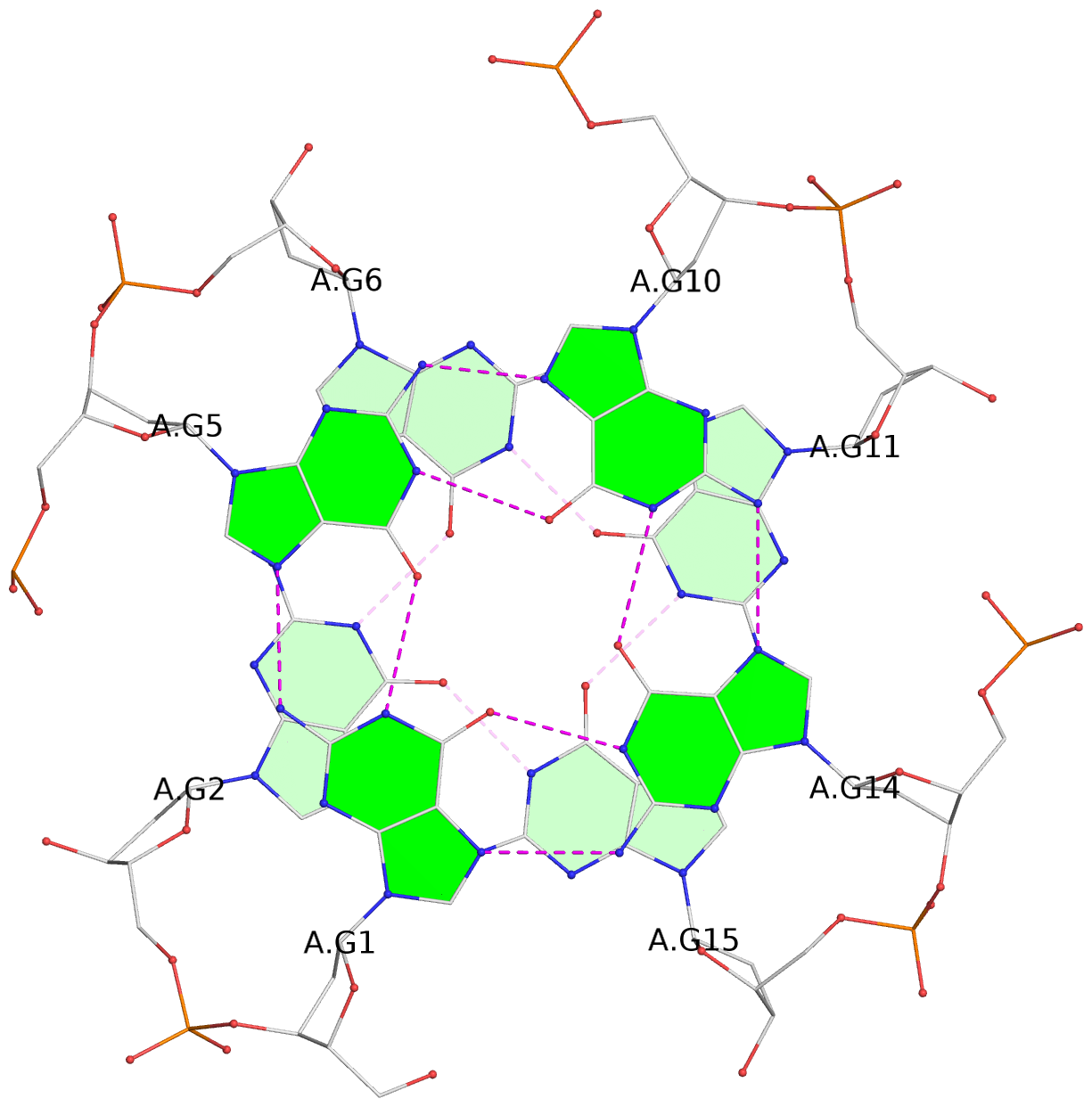
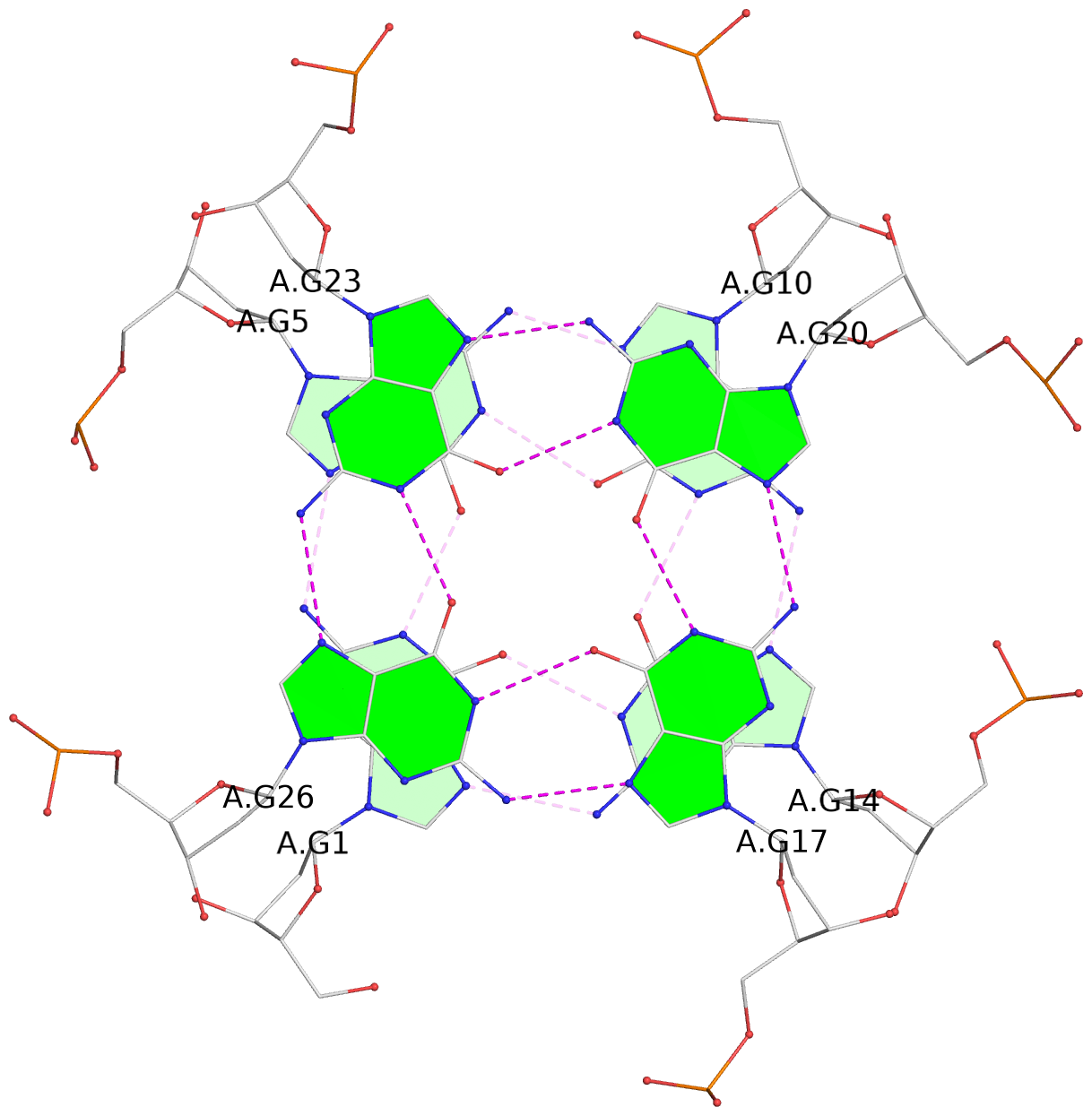
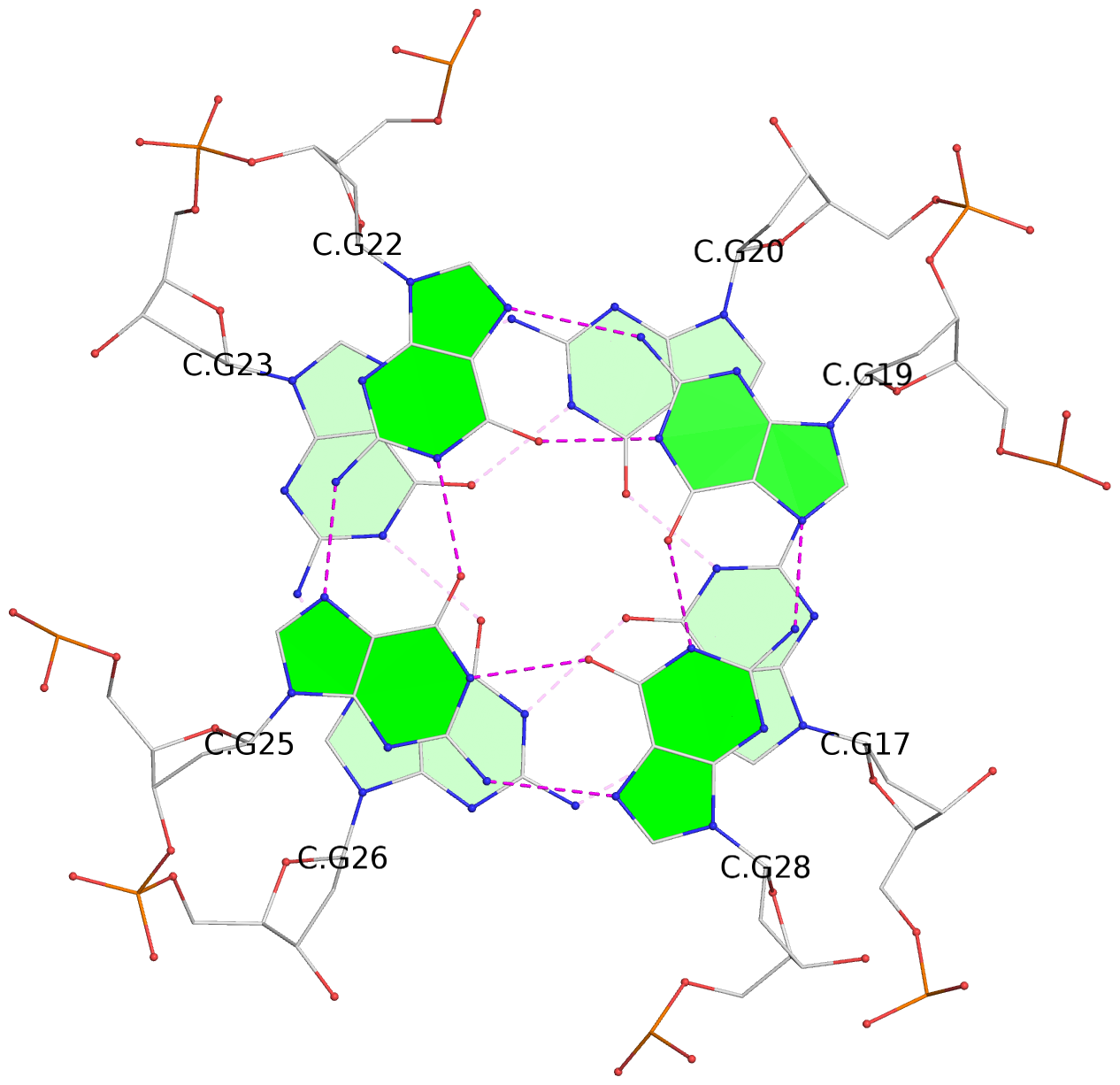
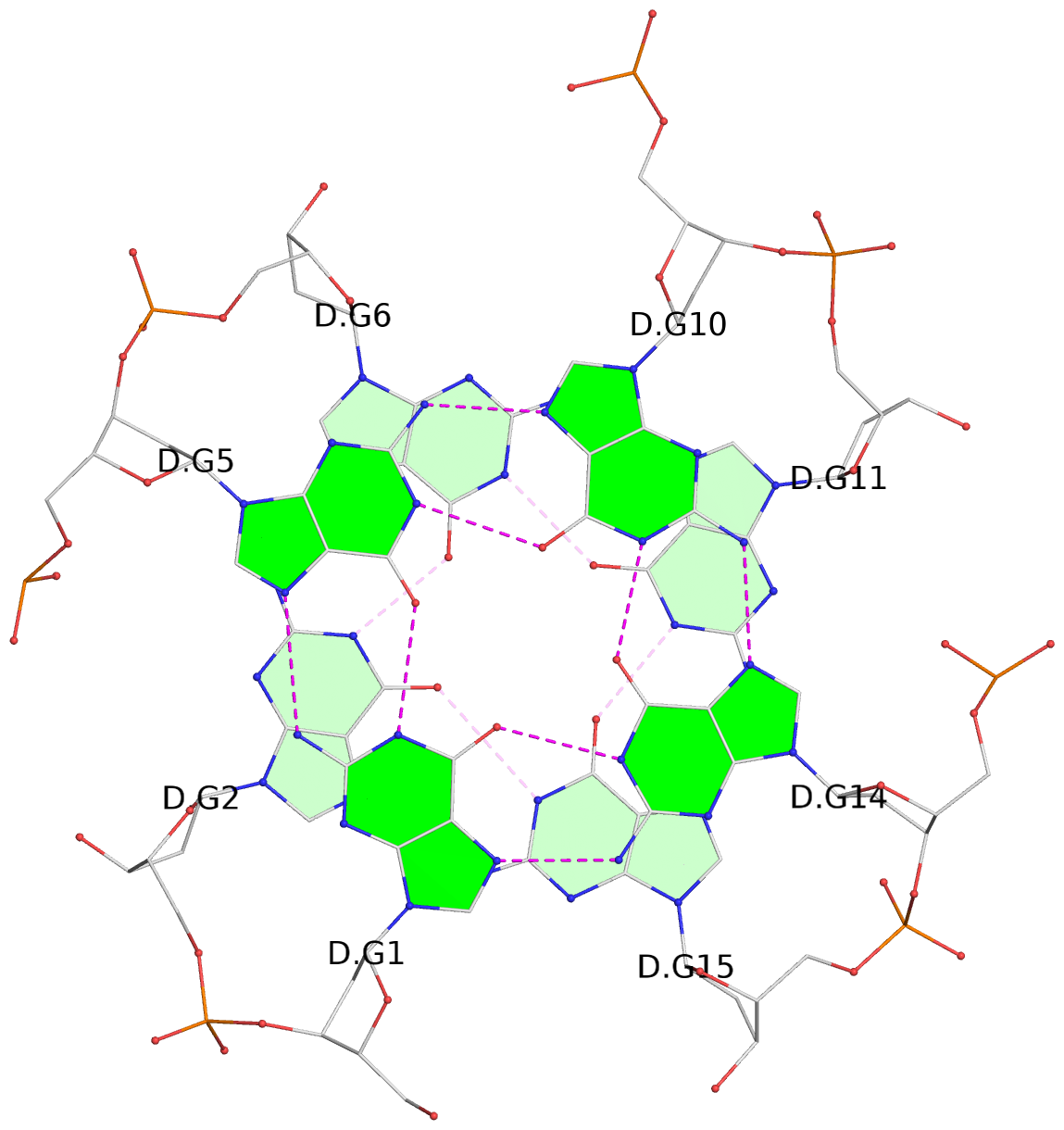
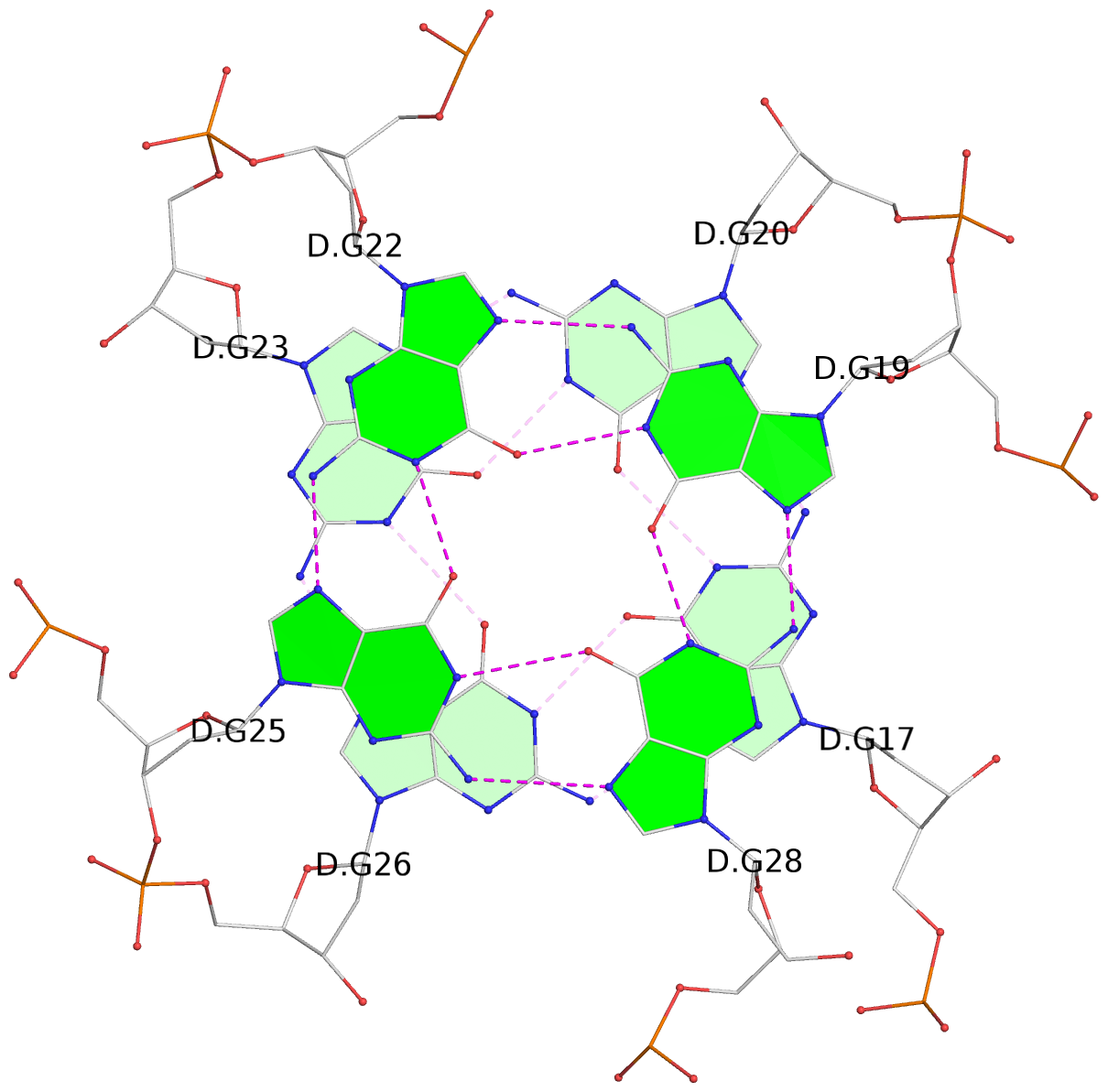
List of 16 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.163 type=other nts=4 GGGG A.DG1,A.DG5,A.DG10,A.DG14 2 glyco-bond=---- sugar=.--- groove=---- planarity=0.156 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG11,A.DG15 3 glyco-bond=---- sugar=---- groove=---- planarity=0.140 type=planar nts=4 GGGG A.DG17,A.DG20,A.DG23,A.DG26 4 glyco-bond=---- sugar=---- groove=---- planarity=0.239 type=other nts=4 GGGG A.DG19,A.DG22,A.DG25,A.DG28 5 glyco-bond=---- sugar=---- groove=---- planarity=0.203 type=other nts=4 GGGG B.DG1,B.DG5,B.DG10,B.DG14 6 glyco-bond=---- sugar=---- groove=---- planarity=0.294 type=other nts=4 GGGG B.DG2,B.DG6,B.DG11,B.DG15 7 glyco-bond=---- sugar=---- groove=---- planarity=0.200 type=other nts=4 GGGG B.DG17,B.DG20,B.DG23,B.DG26 8 glyco-bond=---- sugar=---- groove=---- planarity=0.305 type=other nts=4 GGGG B.DG19,B.DG22,B.DG25,B.DG28 9 glyco-bond=---- sugar=---- groove=---- planarity=0.156 type=planar nts=4 GGGG C.DG1,C.DG5,C.DG10,C.DG14 10 glyco-bond=---- sugar=---- groove=---- planarity=0.204 type=other nts=4 GGGG C.DG2,C.DG6,C.DG11,C.DG15 11 glyco-bond=---- sugar=---- groove=---- planarity=0.132 type=planar nts=4 GGGG C.DG17,C.DG20,C.DG23,C.DG26 12 glyco-bond=---- sugar=---- groove=---- planarity=0.297 type=other nts=4 GGGG C.DG19,C.DG22,C.DG25,C.DG28 13 glyco-bond=---- sugar=---- groove=---- planarity=0.096 type=planar nts=4 GGGG D.DG1,D.DG5,D.DG10,D.DG14 14 glyco-bond=---- sugar=---- groove=---- planarity=0.169 type=other nts=4 GGGG D.DG2,D.DG6,D.DG11,D.DG15 15 glyco-bond=---- sugar=---- groove=---- planarity=0.158 type=planar nts=4 GGGG D.DG17,D.DG20,D.DG23,D.DG26 16 glyco-bond=---- sugar=---- groove=---- planarity=0.261 type=other nts=4 GGGG D.DG19,D.DG22,D.DG25,D.DG28
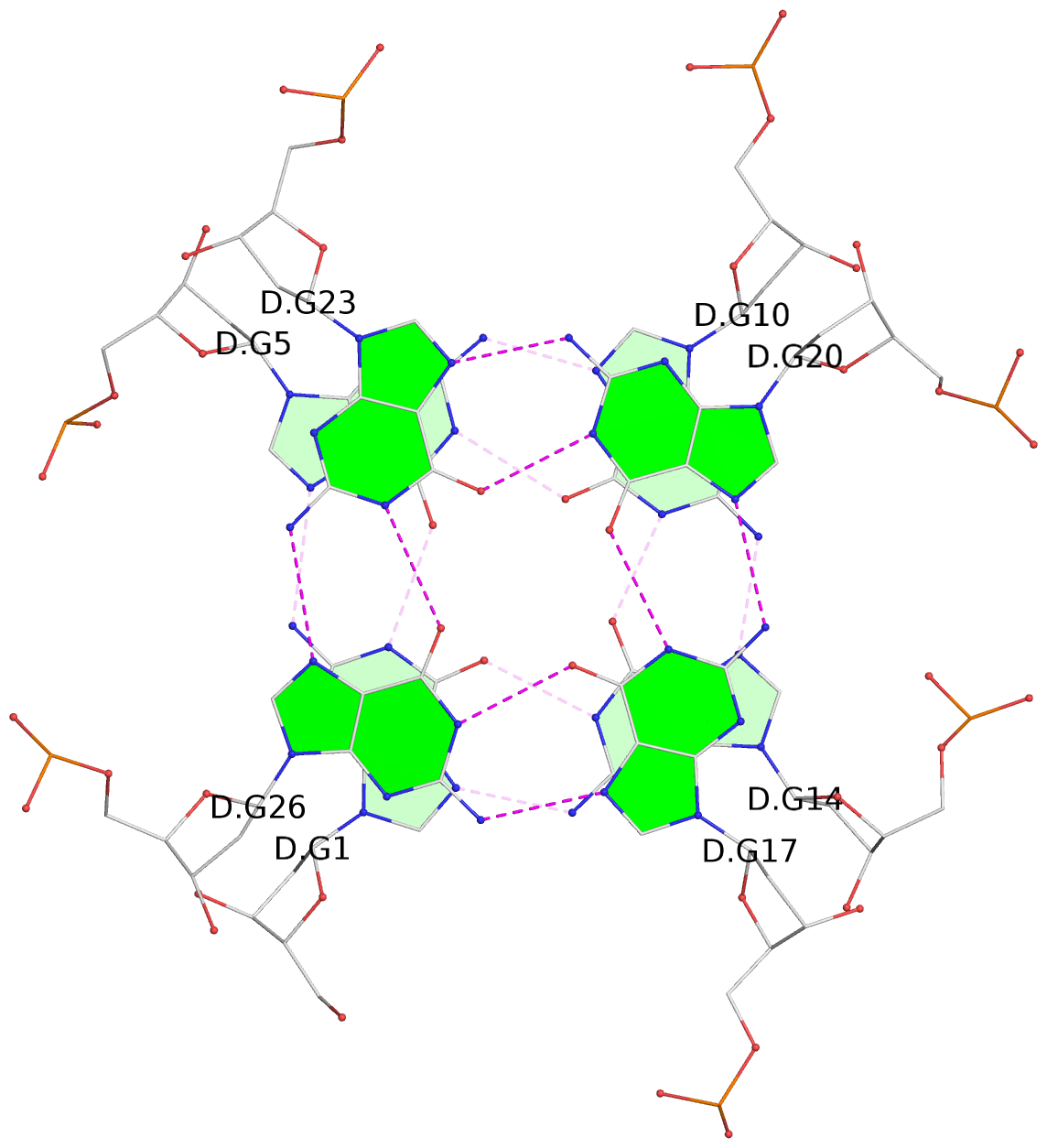
List of 4 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#3, 4 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#4, 4 G-tetrad layers, INTRA-molecular, with 1 stem
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#3, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#4, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
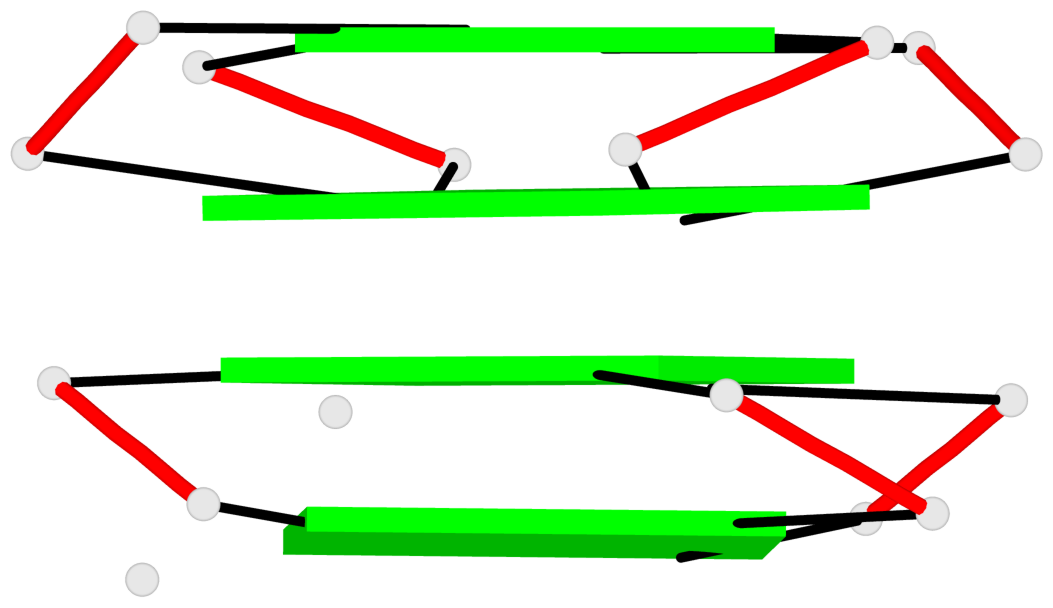
List of 8 non-stem G4-loops (including the two closing Gs)
1 type=V-shaped helix=#1 nts=4 GTGG A.DG17,A.DT18,A.DG19,A.DG20 2* type=V-shaped helix=#1 nts=4 GGTG A.DG25,A.DG26,A.DT27,A.DG28 3 type=V-shaped helix=#2 nts=4 GTGG B.DG17,B.DT18,B.DG19,B.DG20 4* type=V-shaped helix=#2 nts=4 GGTG B.DG25,B.DG26,B.DT27,B.DG28 5 type=V-shaped helix=#3 nts=4 GTGG C.DG17,C.DT18,C.DG19,C.DG20 6* type=V-shaped helix=#3 nts=4 GGTG C.DG25,C.DG26,C.DT27,C.DG28 7 type=V-shaped helix=#4 nts=4 GTGG D.DG17,D.DT18,D.DG19,D.DG20 8* type=V-shaped helix=#4 nts=4 GGTG D.DG25,D.DG26,D.DT27,D.DG28