Detailed DSSR results for the G-quadruplex: PDB entry 6xt7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6xt7
- Class
- DNA
- Method
- X-ray (1.56 Å)
- Summary
- Tel25 hybrid four-quartet g-quadruplex with k+
- Reference
- Li K, Yatsunyk L, Neidle S (2021): "Water spines and networks in G-quadruplex structures." Nucleic Acids Res., 49, 519-528. doi: 10.1093/nar/gkaa1177.
- Abstract
- Quadruplex DNAs can fold into a variety of distinct topologies, depending in part on loop types and orientations of individual strands, as shown by high-resolution crystal and NMR structures. Crystal structures also show associated water molecules. We report here on an analysis of the hydration arrangements around selected folded quadruplex DNAs, which has revealed several prominent features that re-occur in related structures. Many of the primary-sphere water molecules are found in the grooves and loop regions of these structures. At least one groove in anti-parallel and hybrid quadruplex structures is long and narrow and contains an extensive spine of linked primary-sphere water molecules. This spine is analogous to but fundamentally distinct from the well-characterized spine observed in the minor groove of A/T-rich duplex DNA, in that every water molecule in the continuous quadruplex spines makes a direct hydrogen bond contact with groove atoms, principally phosphate oxygen atoms lining groove walls and guanine base nitrogen atoms on the groove floor. By contrast, parallel quadruplexes do not have extended grooves, but primary-sphere water molecules still cluster in them and are especially associated with the loops, helping to stabilize loop conformations.
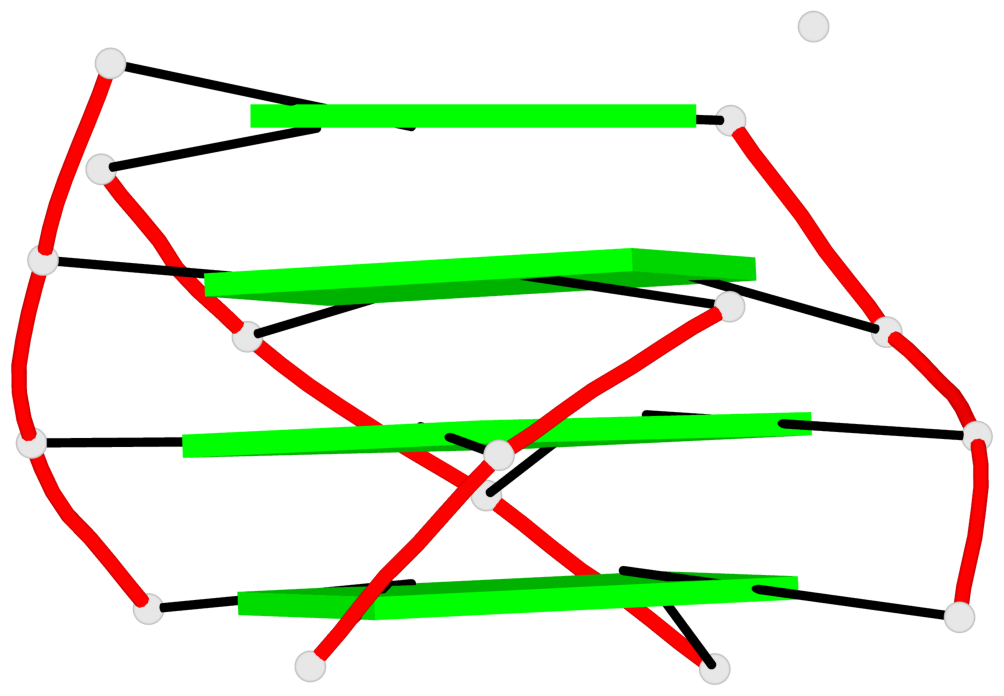
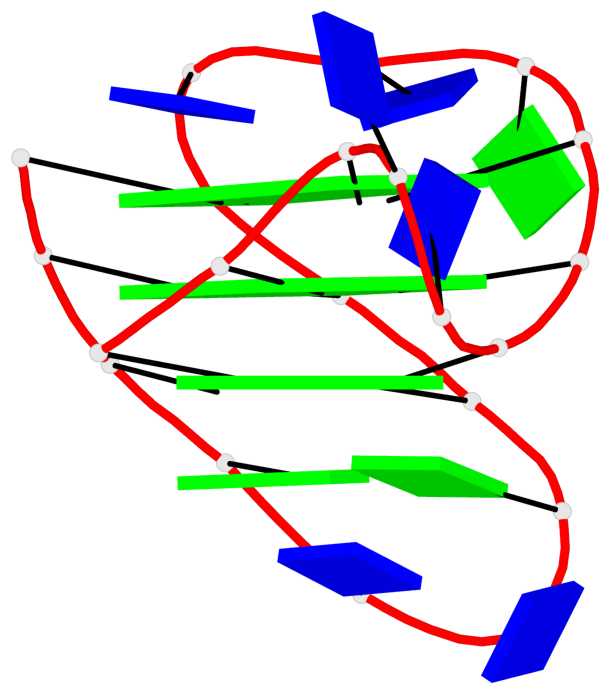
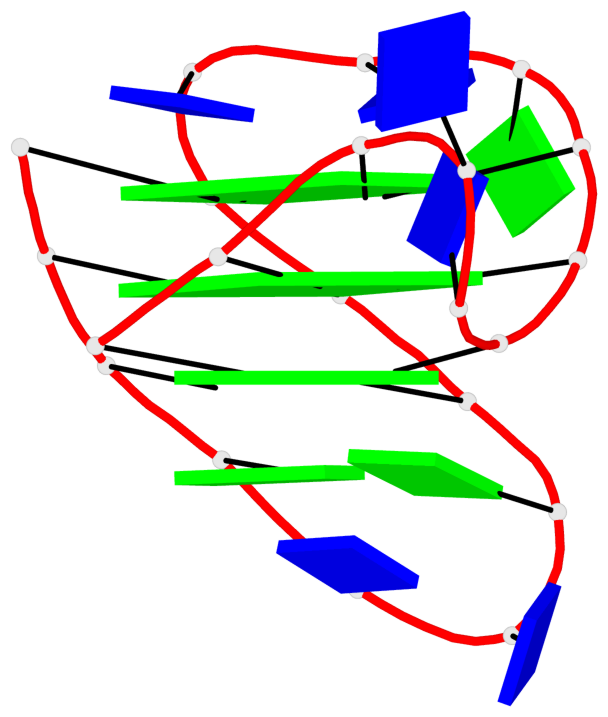
- G4 notes
- 16 G-tetrads, 4 G4 helices, 4 G4 stems, 3(-P-Lw-Ln), hybrid-1(3+1), UUDU
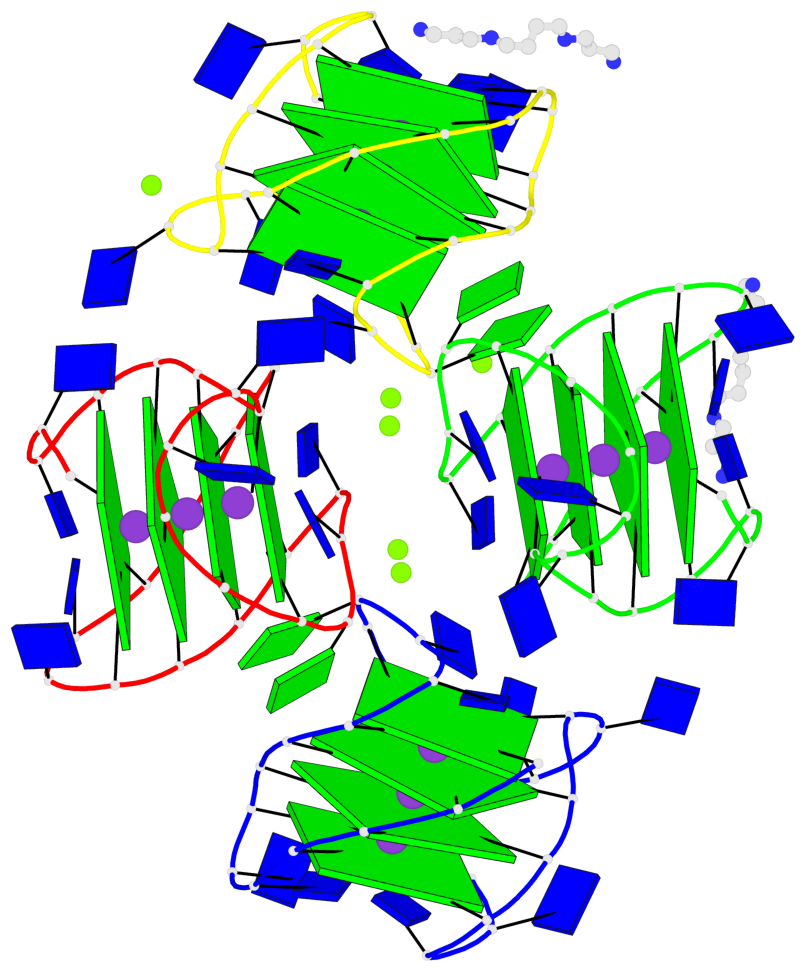
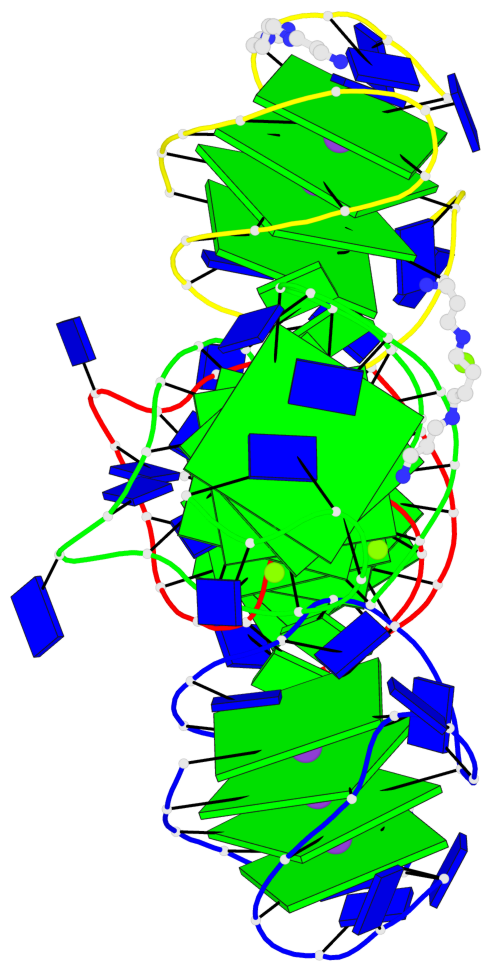
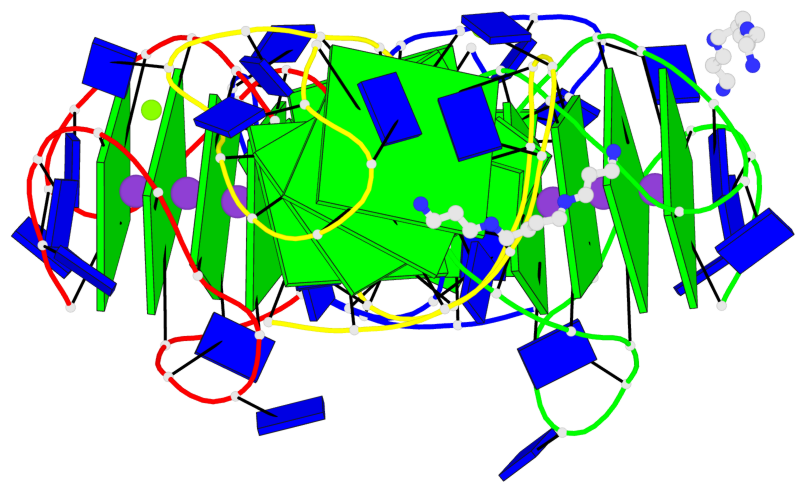
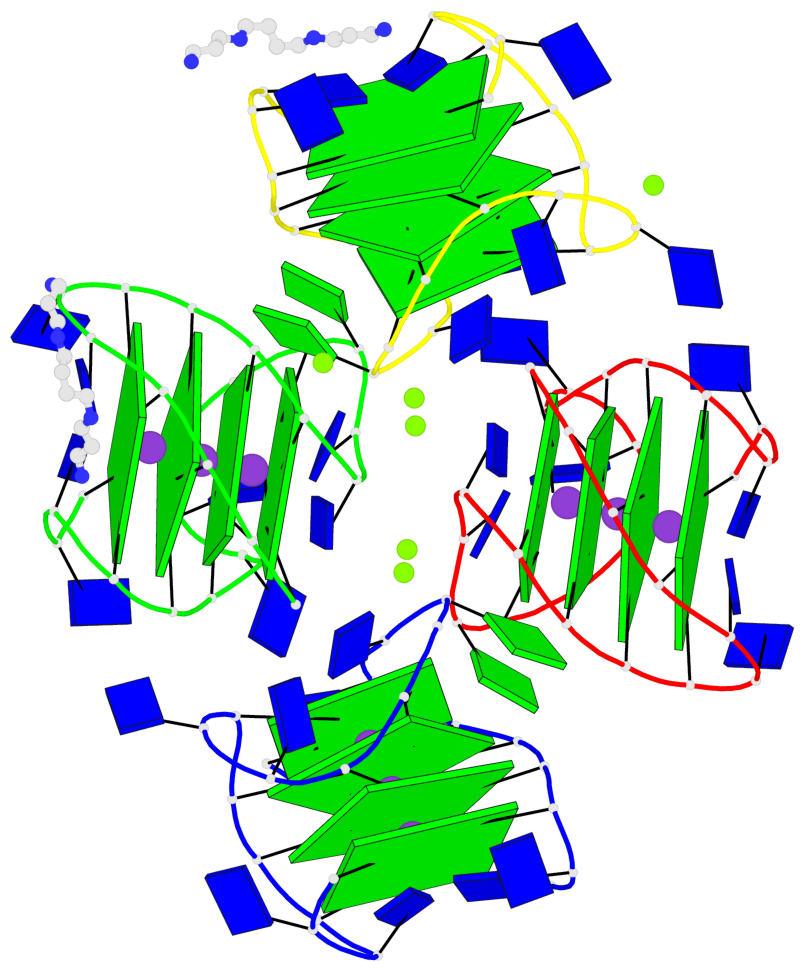
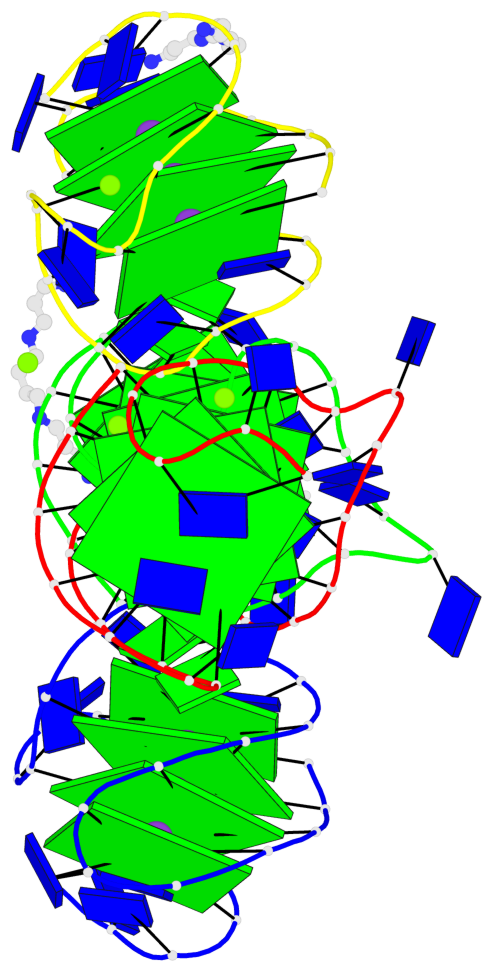
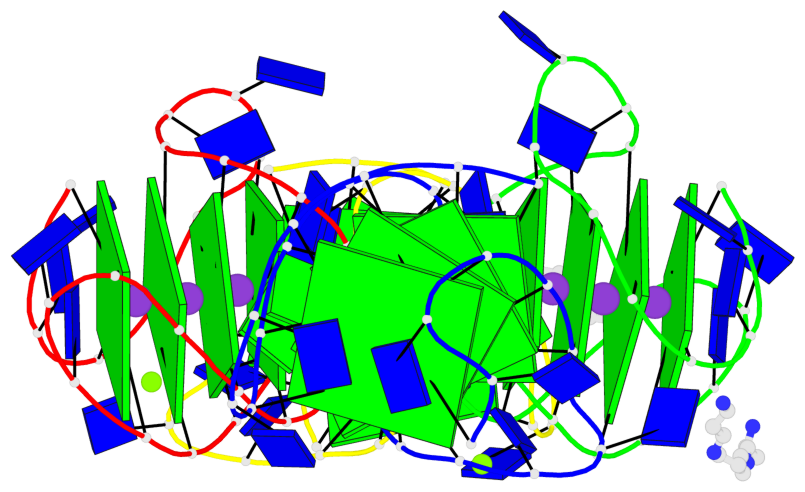
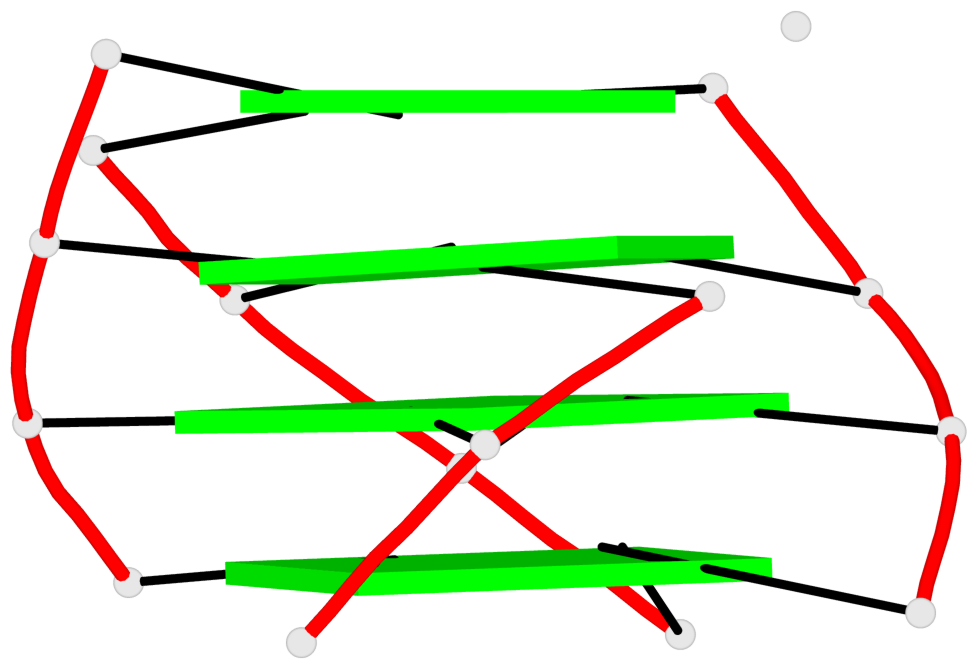
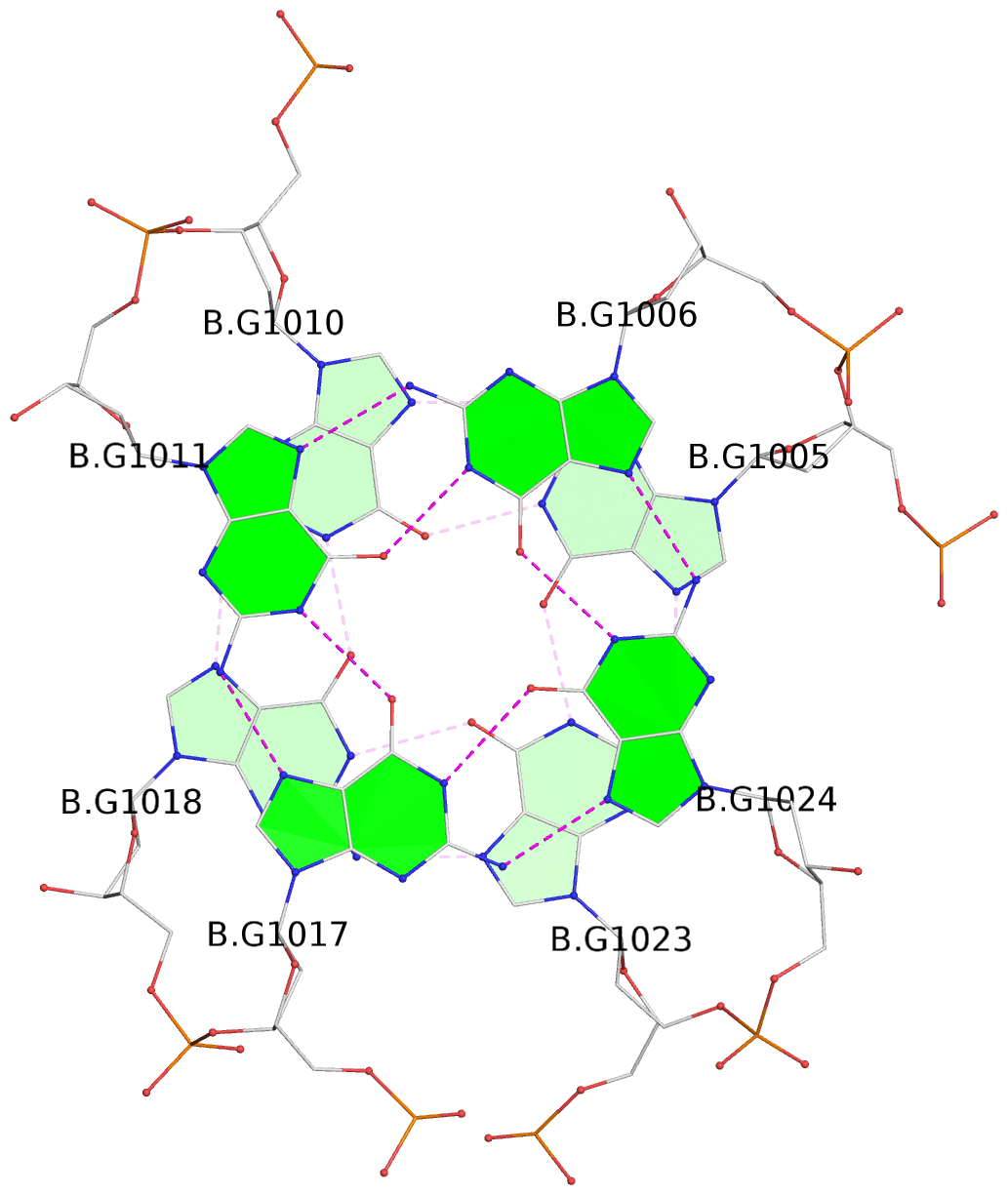
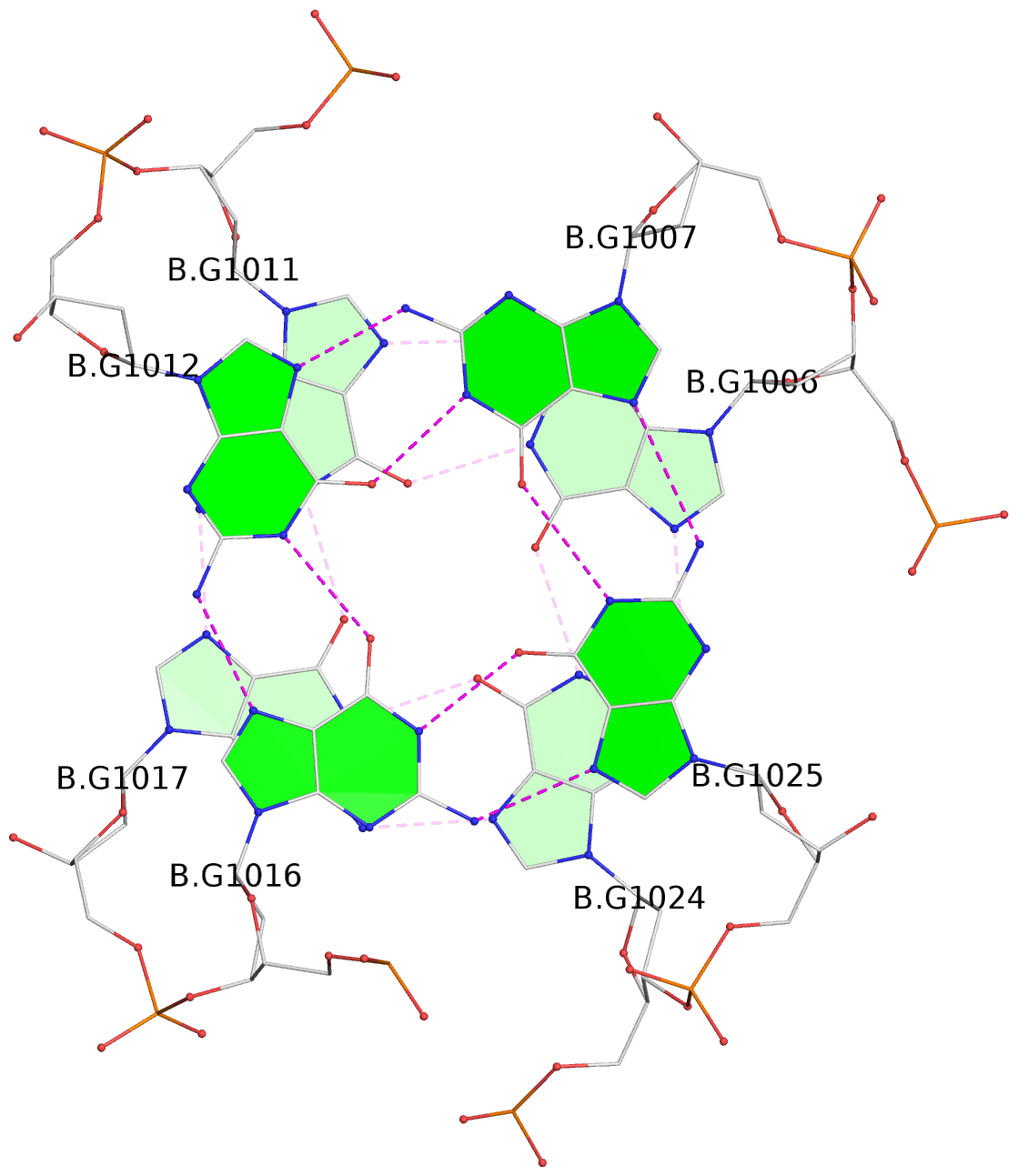
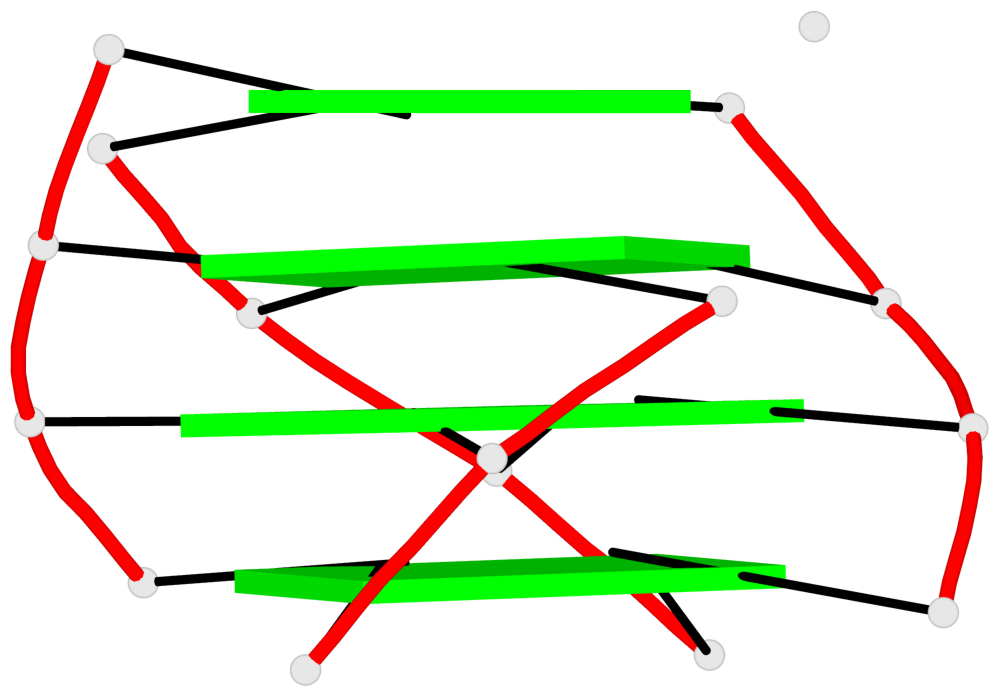
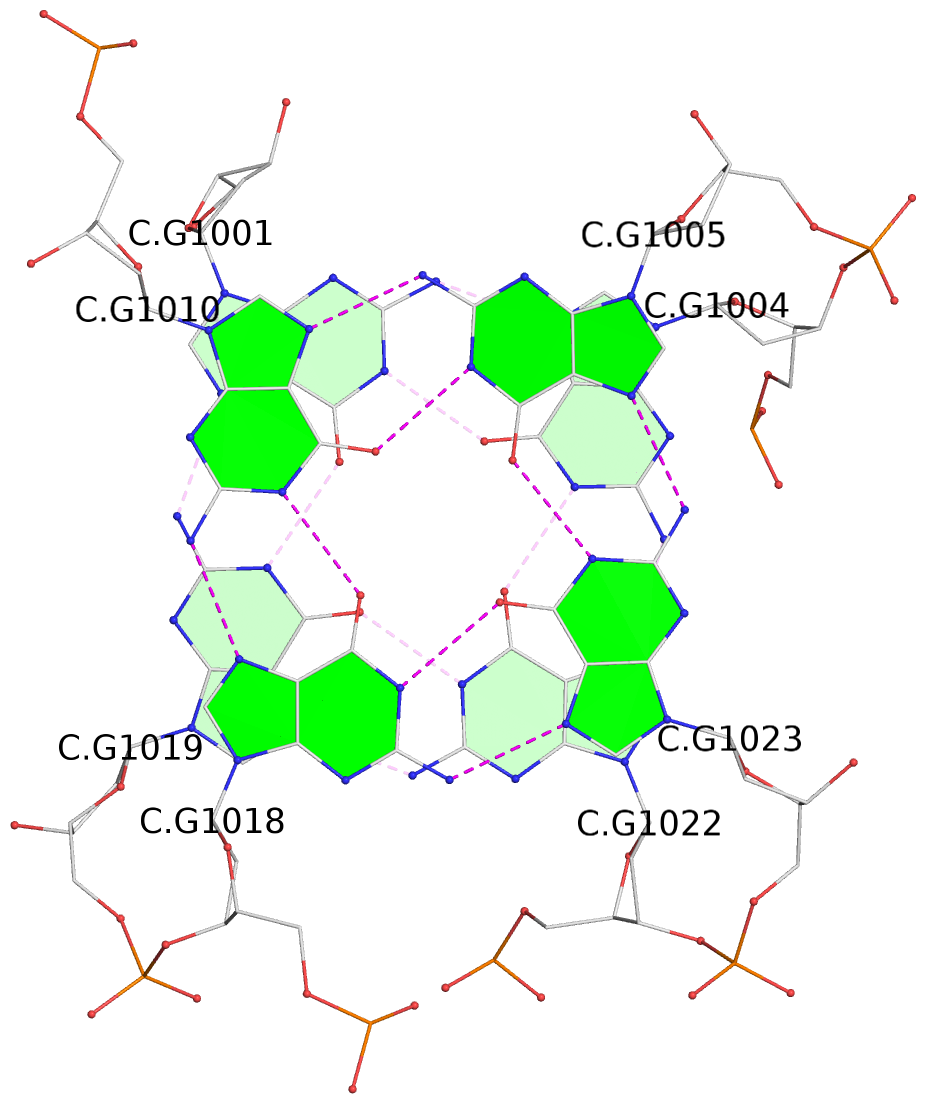
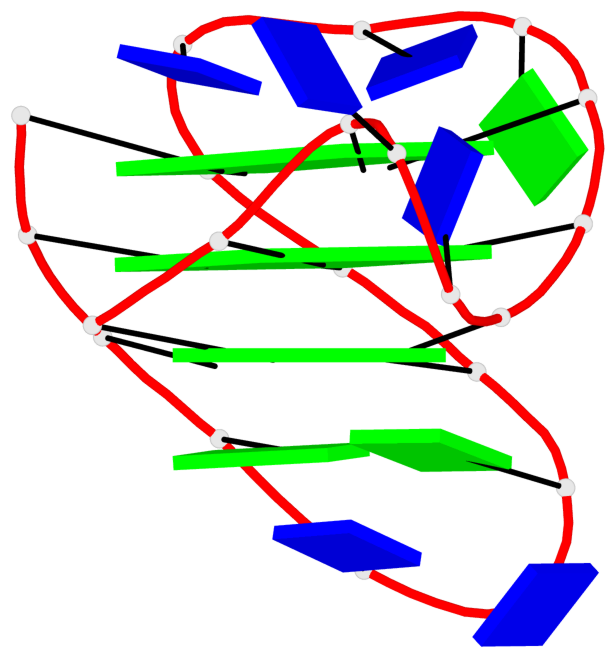
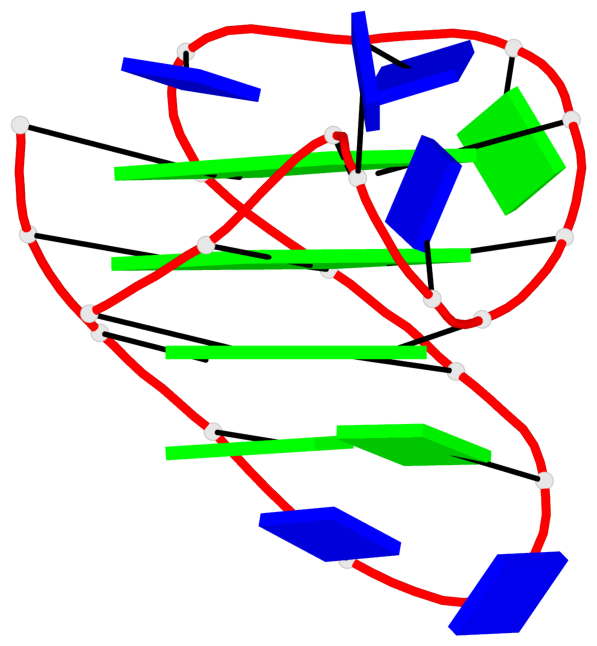
Base-block schematics in six views
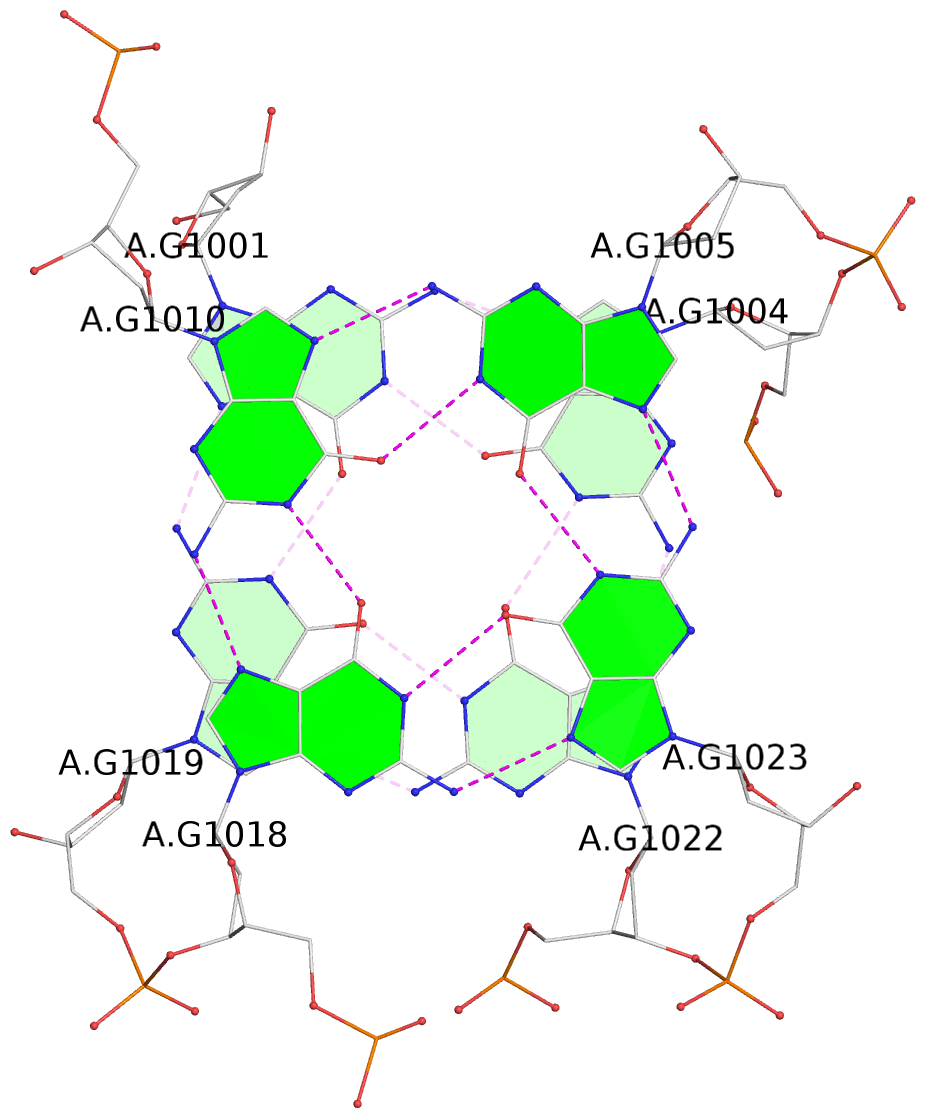
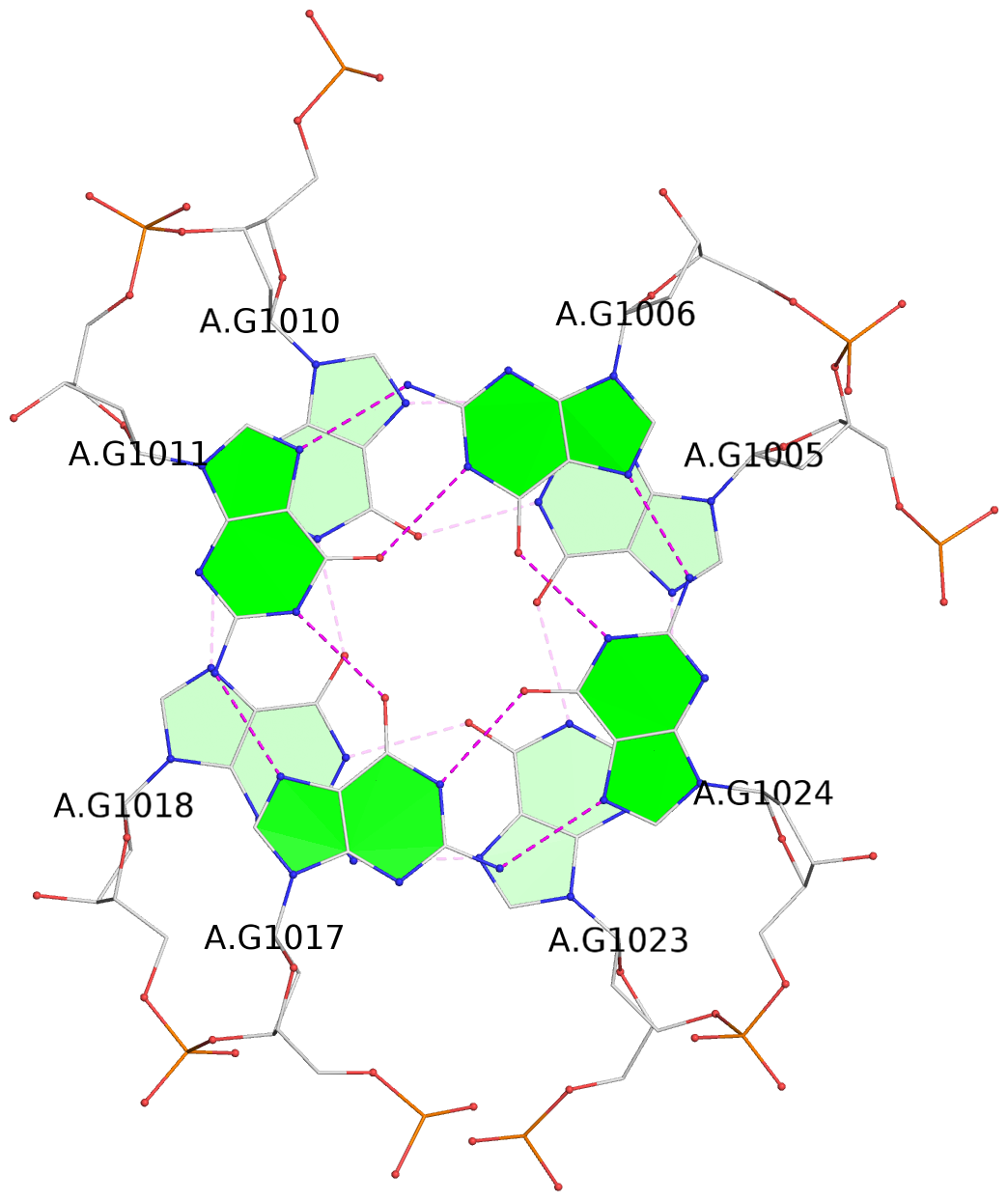
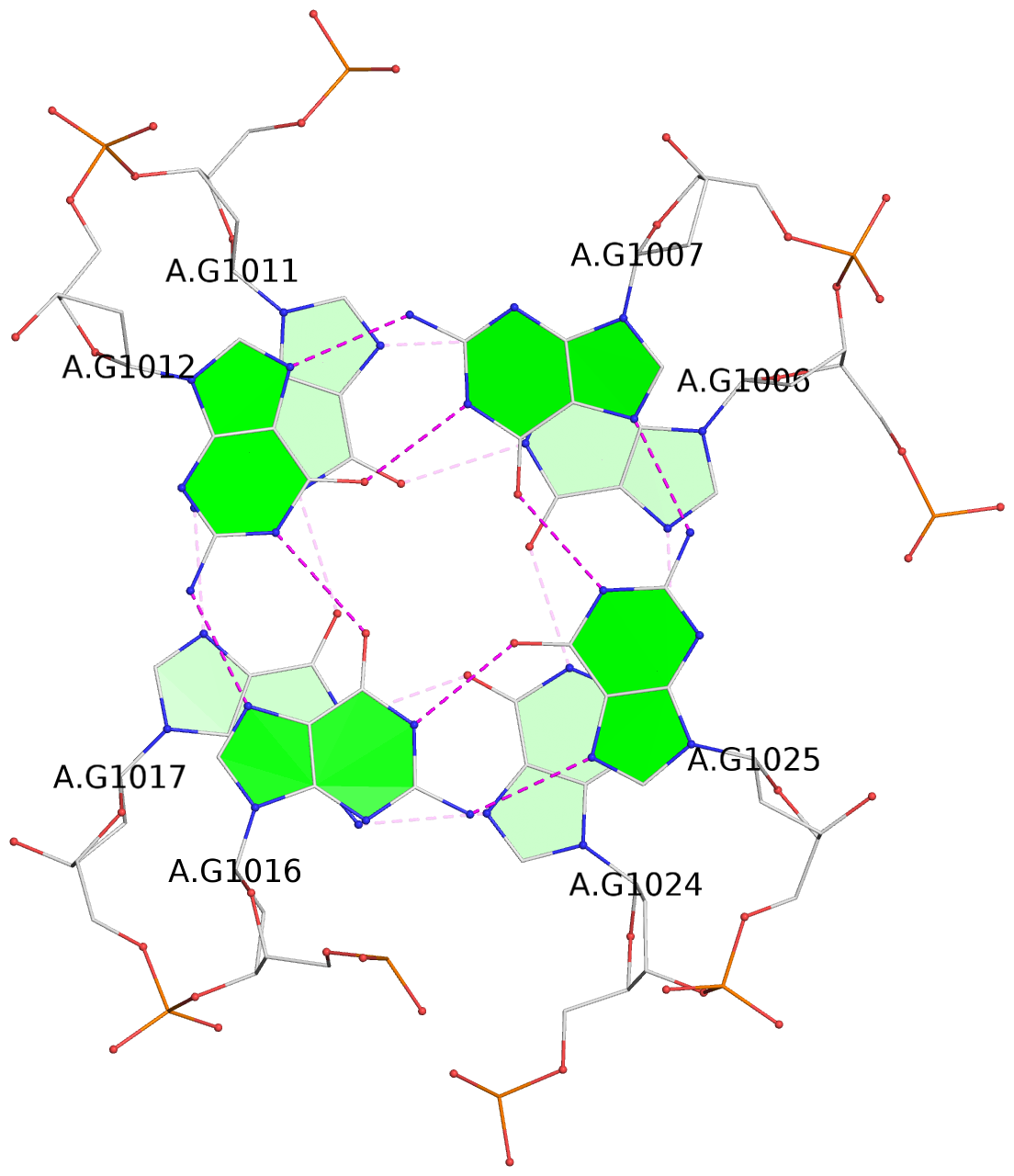
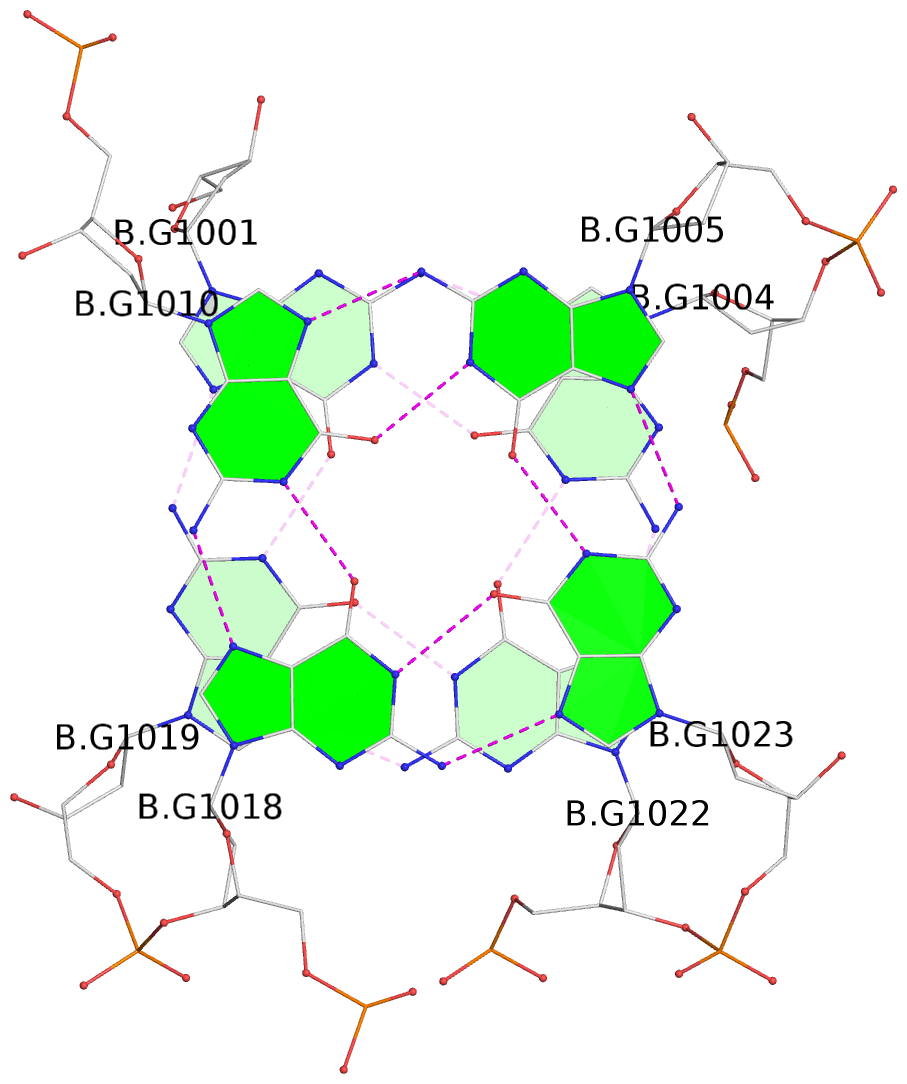
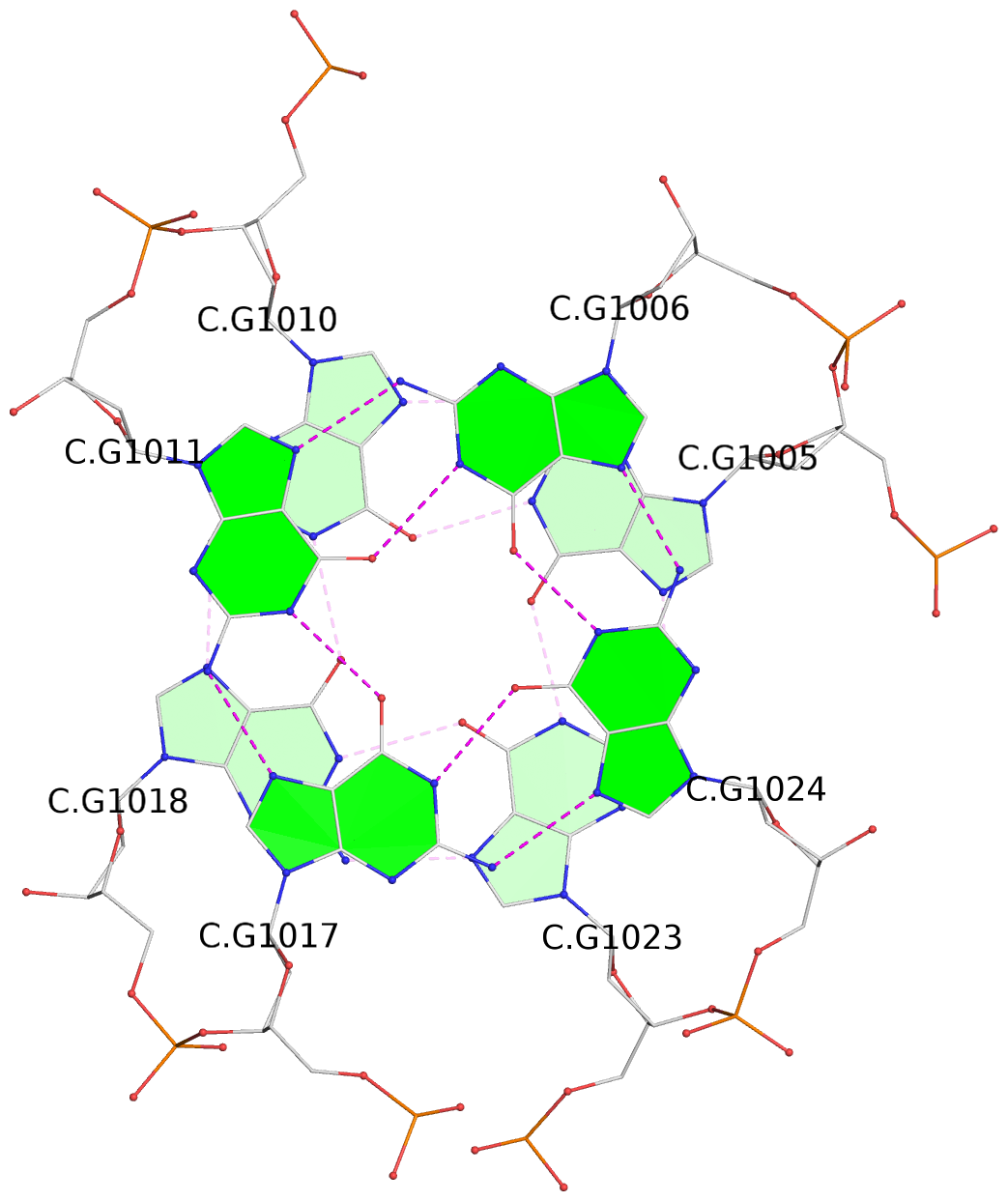
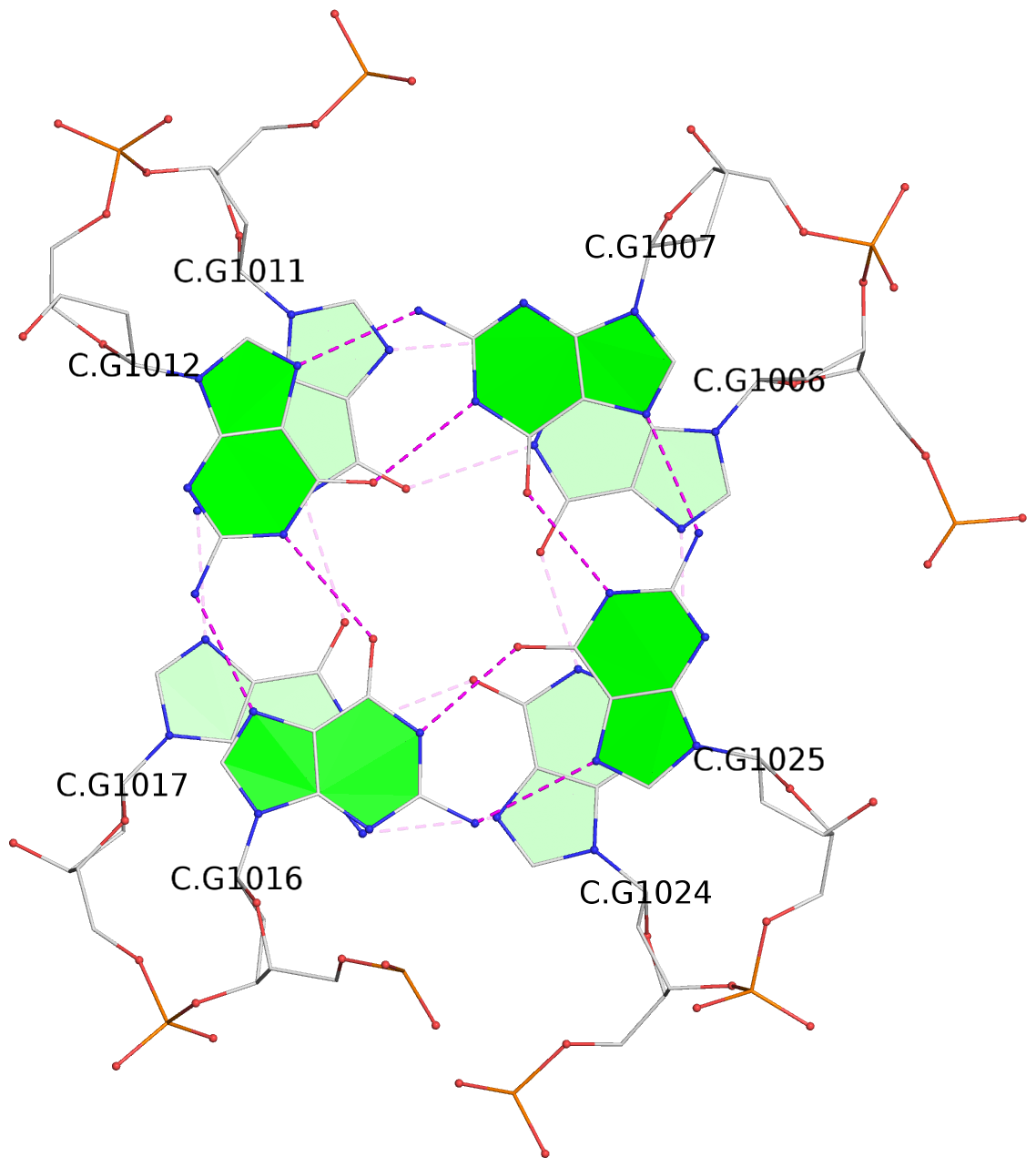
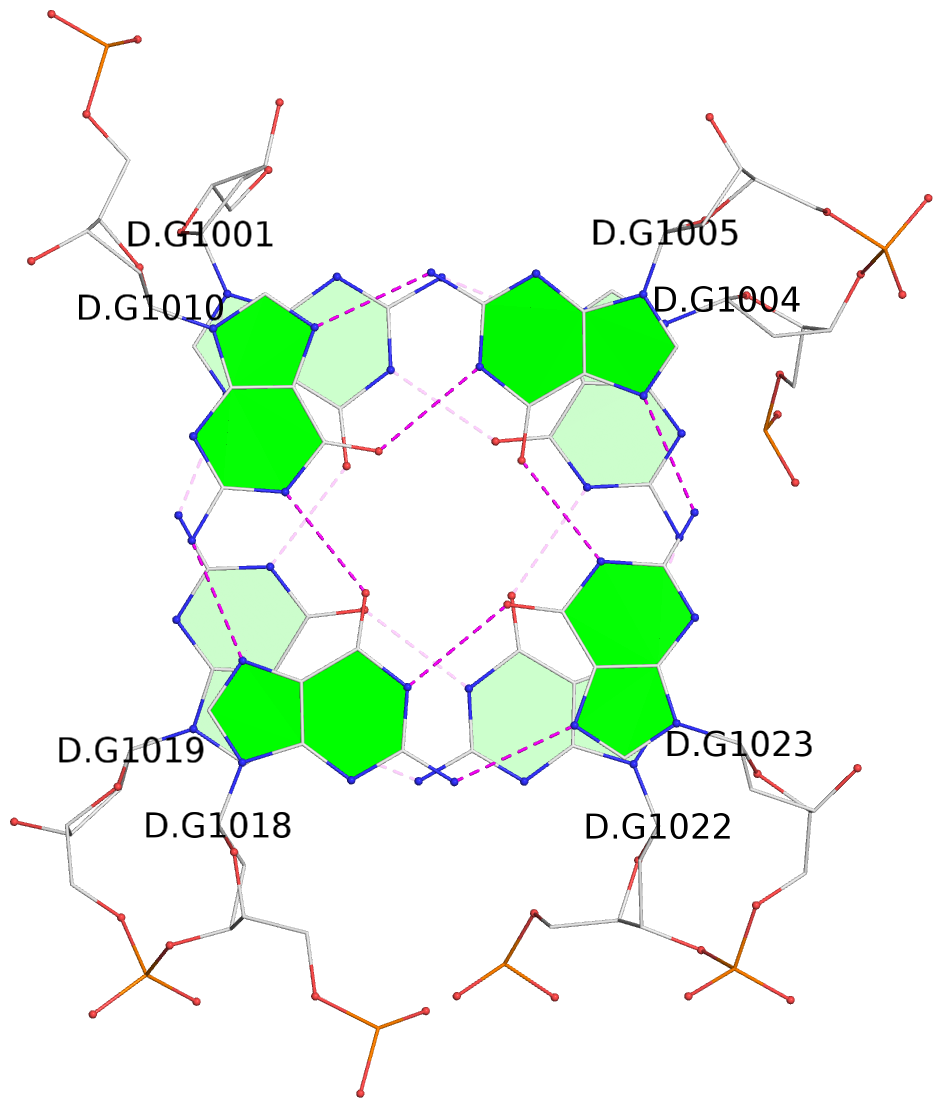
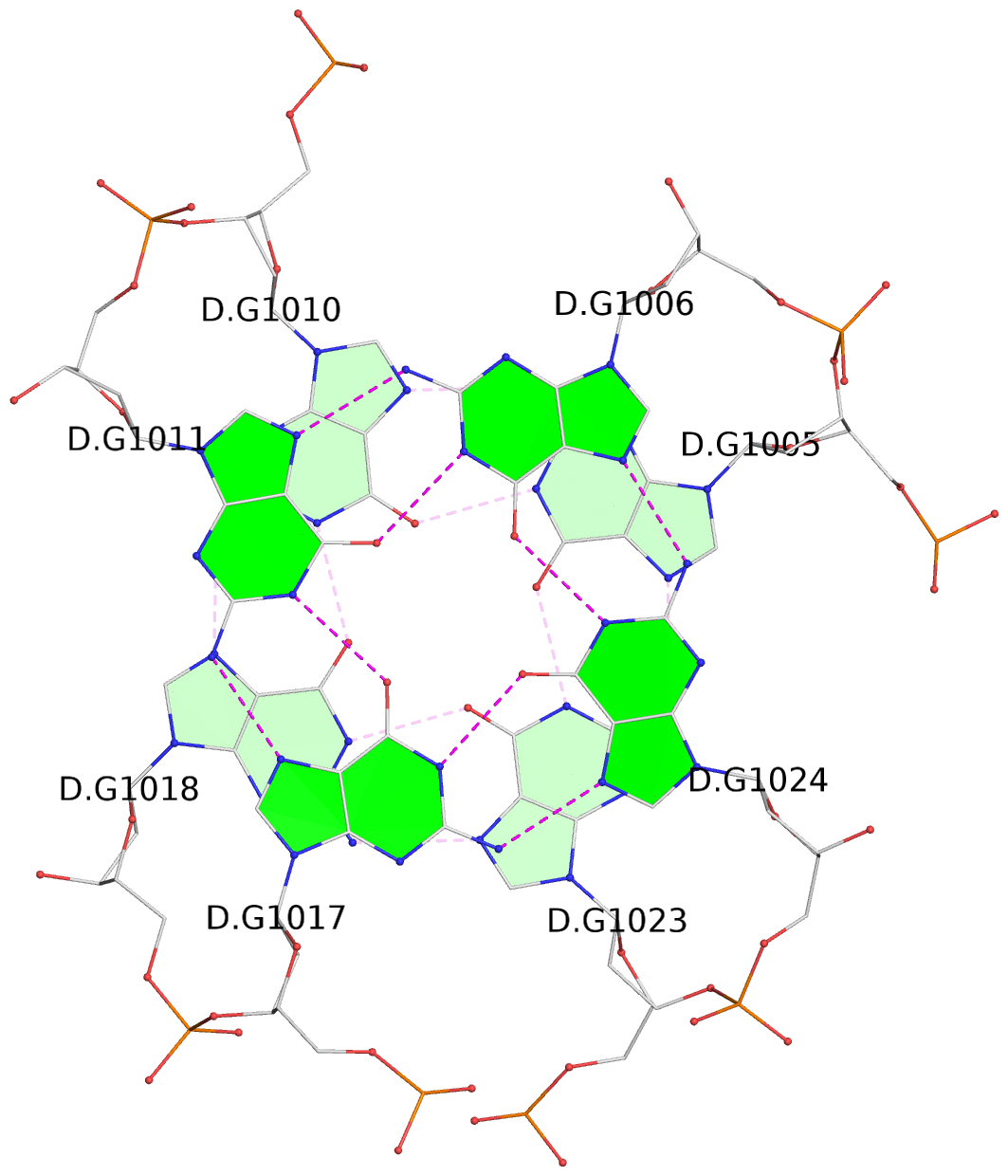
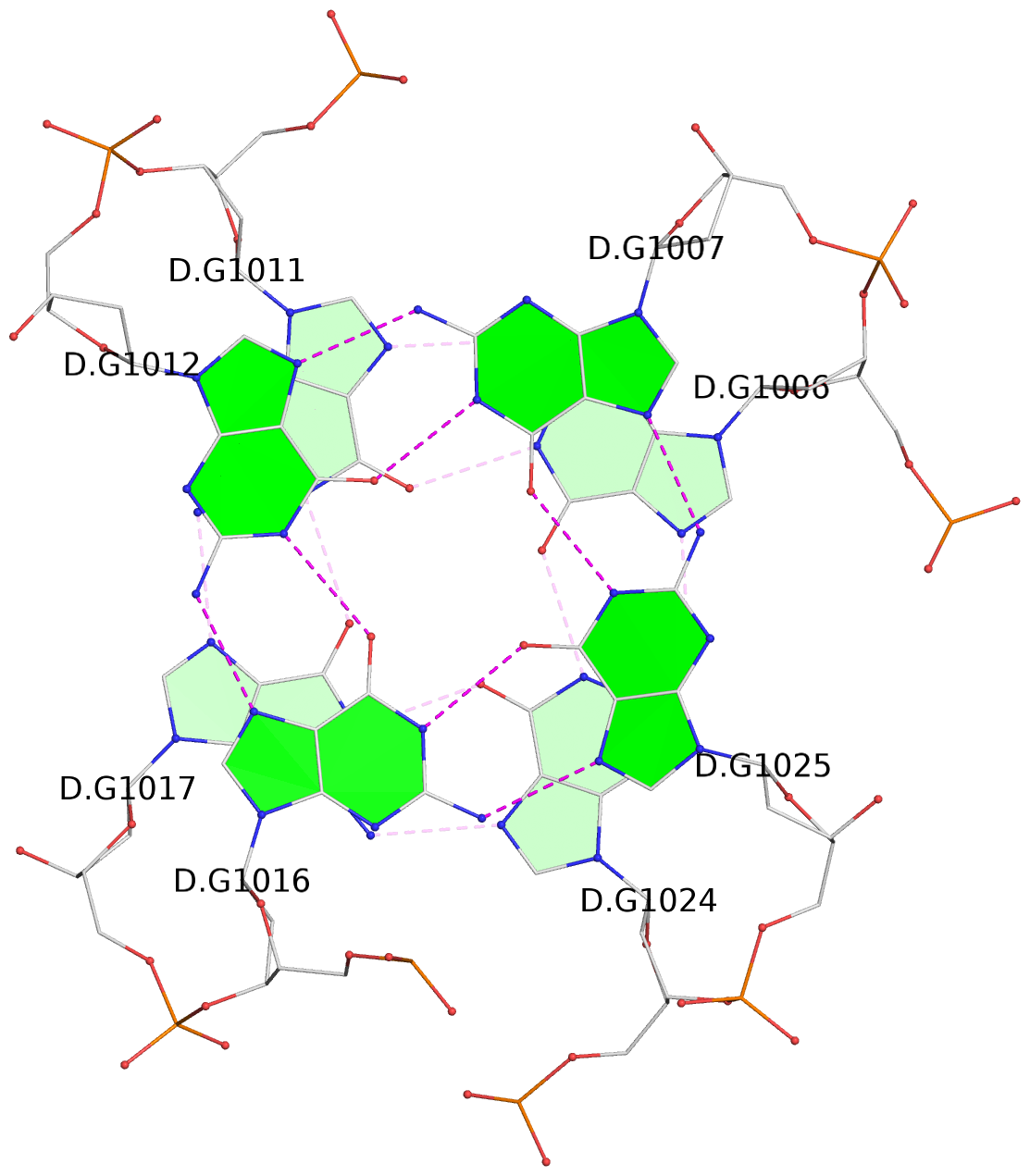
List of 16 G-tetrads
1 glyco-bond=s-ss sugar=3--- groove=wn-- planarity=0.202 type=other nts=4 GGGG A.DG1001,A.DG1019,A.DG1022,A.DG1004 2 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.172 type=other nts=4 GGGG A.DG1005,A.DG1010,A.DG1018,A.DG1023 3 glyco-bond=--s- sugar=---. groove=-wn- planarity=0.182 type=other nts=4 GGGG A.DG1006,A.DG1011,A.DG1017,A.DG1024 4 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.353 type=bowl nts=4 GGGG A.DG1007,A.DG1012,A.DG1016,A.DG1025 5 glyco-bond=s-ss sugar=3--- groove=wn-- planarity=0.222 type=other nts=4 GGGG B.DG1001,B.DG1019,B.DG1022,B.DG1004 6 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.183 type=other nts=4 GGGG B.DG1005,B.DG1010,B.DG1018,B.DG1023 7 glyco-bond=--s- sugar=---3 groove=-wn- planarity=0.180 type=other nts=4 GGGG B.DG1006,B.DG1011,B.DG1017,B.DG1024 8 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.332 type=bowl nts=4 GGGG B.DG1007,B.DG1012,B.DG1016,B.DG1025 9 glyco-bond=s-ss sugar=3.-- groove=wn-- planarity=0.206 type=other nts=4 GGGG C.DG1001,C.DG1019,C.DG1022,C.DG1004 10 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.188 type=other nts=4 GGGG C.DG1005,C.DG1010,C.DG1018,C.DG1023 11 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.177 type=other nts=4 GGGG C.DG1006,C.DG1011,C.DG1017,C.DG1024 12 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.382 type=bowl nts=4 GGGG C.DG1007,C.DG1012,C.DG1016,C.DG1025 13 glyco-bond=s-ss sugar=3.-- groove=wn-- planarity=0.207 type=other nts=4 GGGG D.DG1001,D.DG1019,D.DG1022,D.DG1004 14 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.164 type=other nts=4 GGGG D.DG1005,D.DG1010,D.DG1018,D.DG1023 15 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.161 type=other nts=4 GGGG D.DG1006,D.DG1011,D.DG1017,D.DG1024 16 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.342 type=bowl nts=4 GGGG D.DG1007,D.DG1012,D.DG1016,D.DG1025
List of 4 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#3, 4 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#4, 4 G-tetrad layers, INTRA-molecular, with 1 stem
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUDU, hybrid-(mixed), 3(-P-Lw-Ln), hybrid-1(3+1)
Stem#2, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUDU, hybrid-(mixed), 3(-P-Lw-Ln), hybrid-1(3+1)
Stem#3, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUDU, hybrid-(mixed), 3(-P-Lw-Ln), hybrid-1(3+1)
Stem#4, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUDU, hybrid-(mixed), 3(-P-Lw-Ln), hybrid-1(3+1)
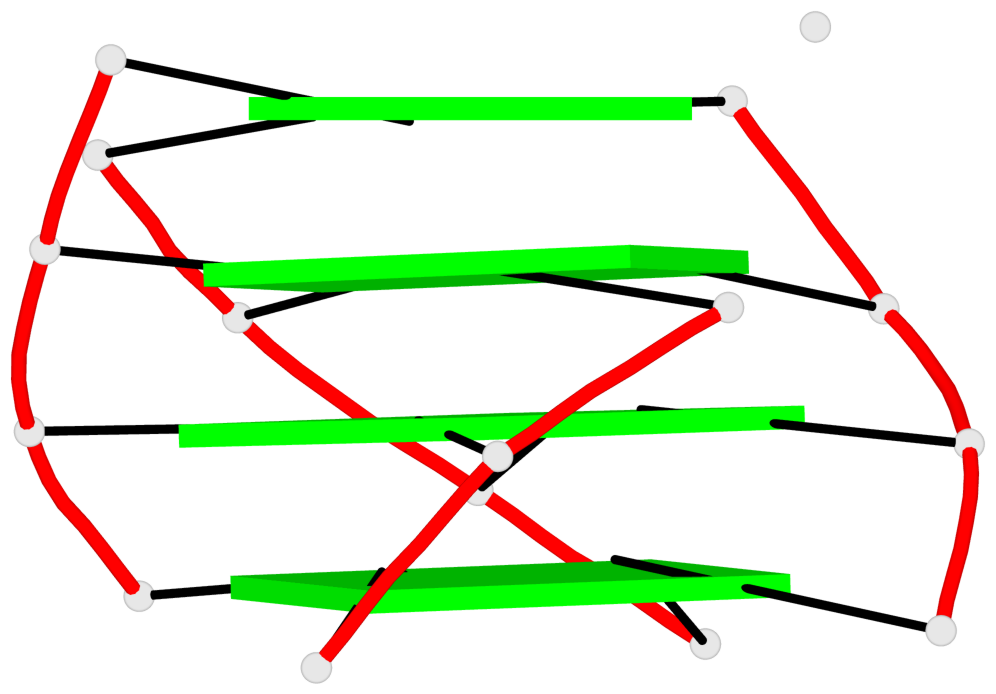
List of 8 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=4 GTTG A.DG1001,A.DT1002,A.DT1003,A.DG1004 2 type=lateral helix=#1 nts=4 GTTG A.DG1019,A.DT1020,A.DT1021,A.DG1022 3 type=lateral helix=#2 nts=4 GTTG B.DG1001,B.DT1002,B.DT1003,B.DG1004 4 type=lateral helix=#2 nts=4 GTTG B.DG1019,B.DT1020,B.DT1021,B.DG1022 5 type=lateral helix=#3 nts=4 GTTG C.DG1001,C.DT1002,C.DT1003,C.DG1004 6 type=lateral helix=#3 nts=4 GTTG C.DG1019,C.DT1020,C.DT1021,C.DG1022 7 type=lateral helix=#4 nts=4 GTTG D.DG1001,D.DT1002,D.DT1003,D.DG1004 8 type=lateral helix=#4 nts=4 GTTG D.DG1019,D.DT1020,D.DT1021,D.DG1022