Detailed DSSR results for the G-quadruplex: PDB entry 6yy4
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6yy4
- Class
- DNA
- Method
- NMR
- Summary
- Parallel 17-mer DNA g-quadruplex
- Reference
- Volek M, Kolesnikova S, Svehlova K, Srb P, Sgallova R, Streckerova T, Redondo JA, Veverka V, Curtis EA (2021): "Overlapping but distinct: a new model for G-quadruplex biochemical specificity." Nucleic Acids Res., 49, 1816-1827. doi: 10.1093/nar/gkab037.
- Abstract
- G-quadruplexes are noncanonical nucleic acid structures formed by stacked guanine tetrads. They are capable of a range of functions and thought to play widespread biological roles. This diversity raises an important question: what determines the biochemical specificity of G-quadruplex structures? The answer is particularly important from the perspective of biological regulation because genomes can contain hundreds of thousands of G-quadruplexes with a range of functions. Here we analyze the specificity of each sequence in a 496-member library of variants of a reference G-quadruplex with respect to five functions. Our analysis shows that the sequence requirements of G-quadruplexes with these functions are different from one another, with some mutations altering biochemical specificity by orders of magnitude. Mutations in tetrads have larger effects than mutations in loops, and changes in specificity are correlated with changes in multimeric state. To complement our biochemical data we determined the solution structure of a monomeric G-quadruplex from the library. The stacked and accessible tetrads rationalize why monomers tend to promote a model peroxidase reaction and generate fluorescence. Our experiments support a model in which the sequence requirements of G-quadruplexes with different functions are overlapping but distinct. This has implications for biological regulation, bioinformatics, and drug design.
- G4 notes
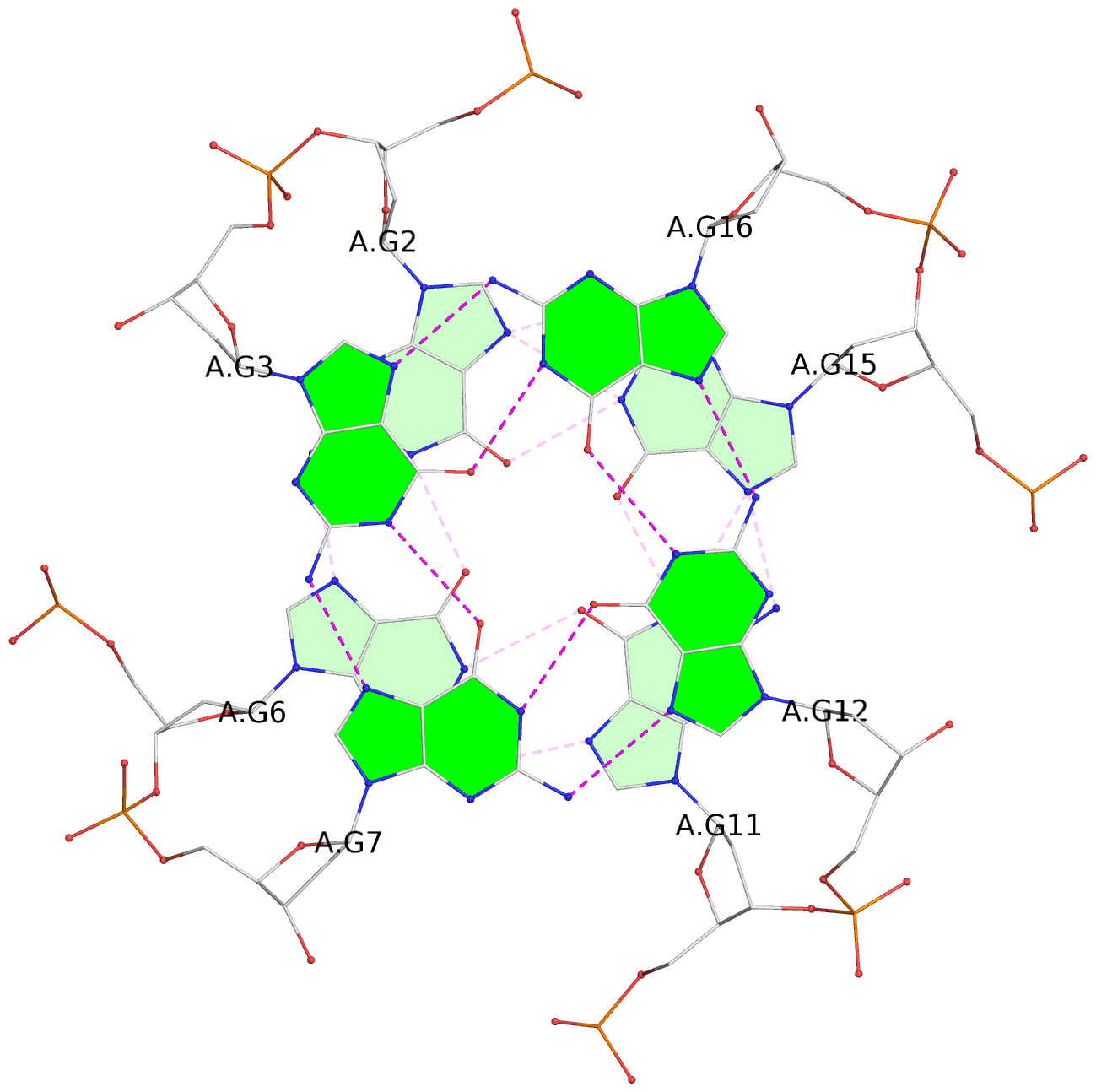
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
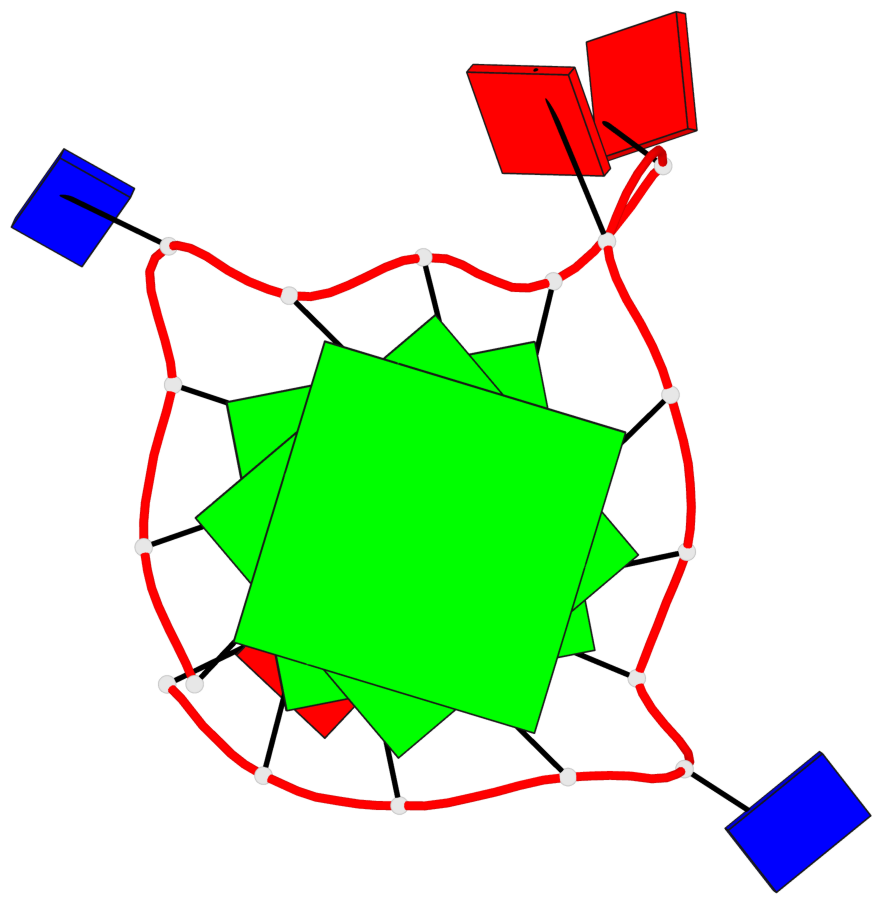
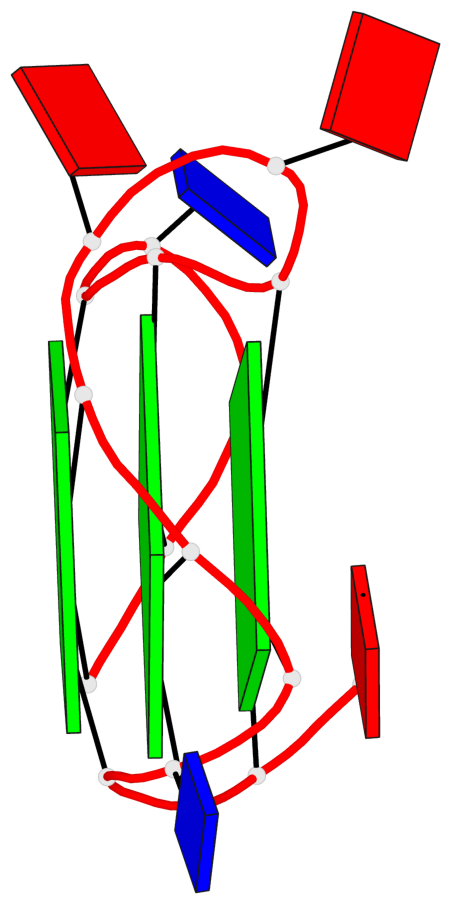
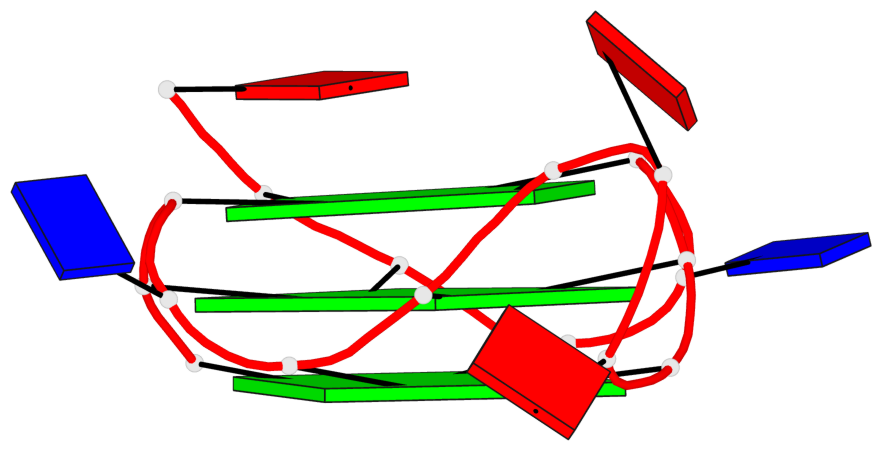
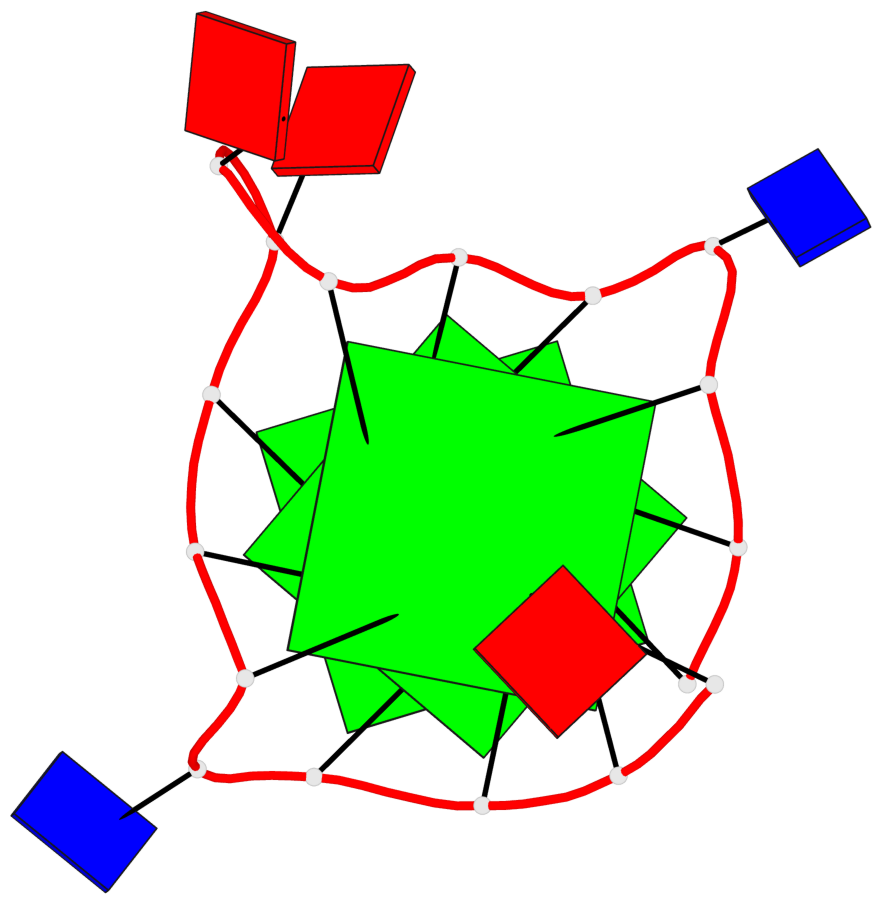
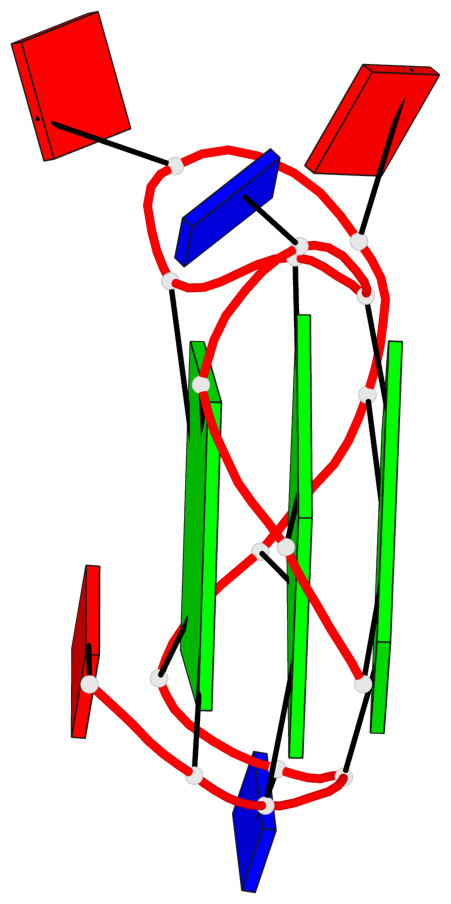
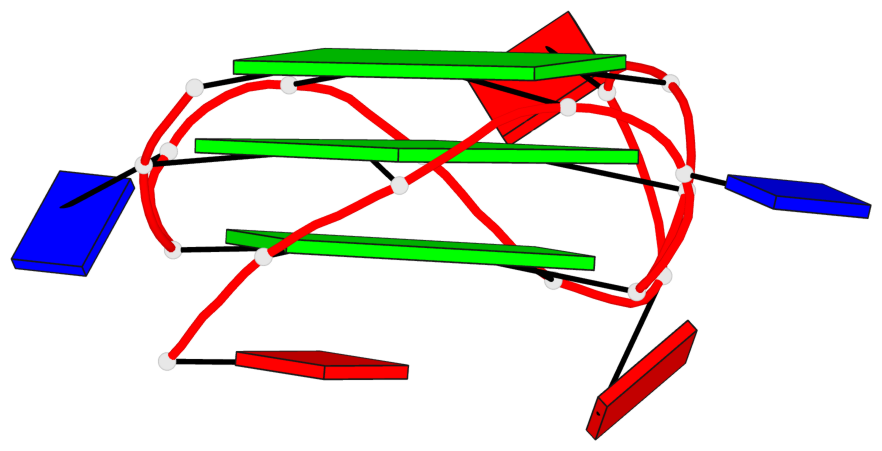
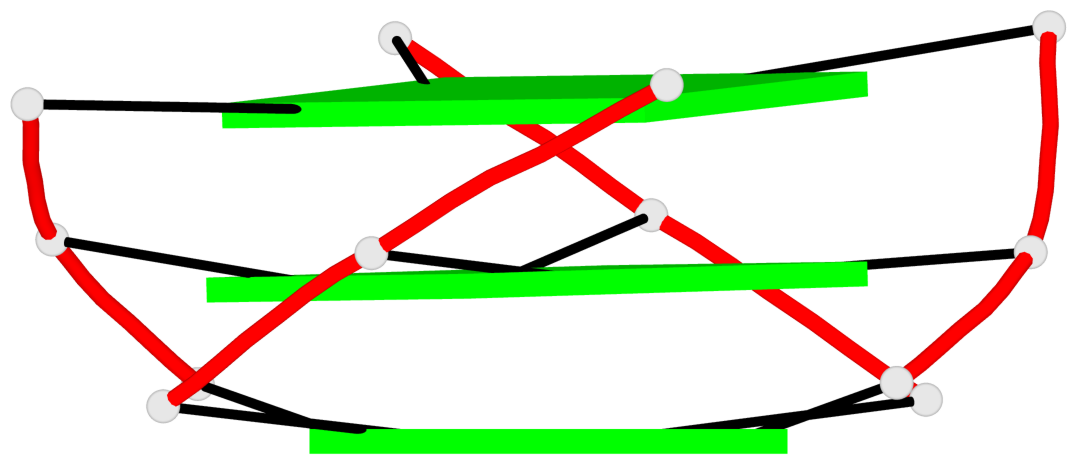
Base-block schematics in six views
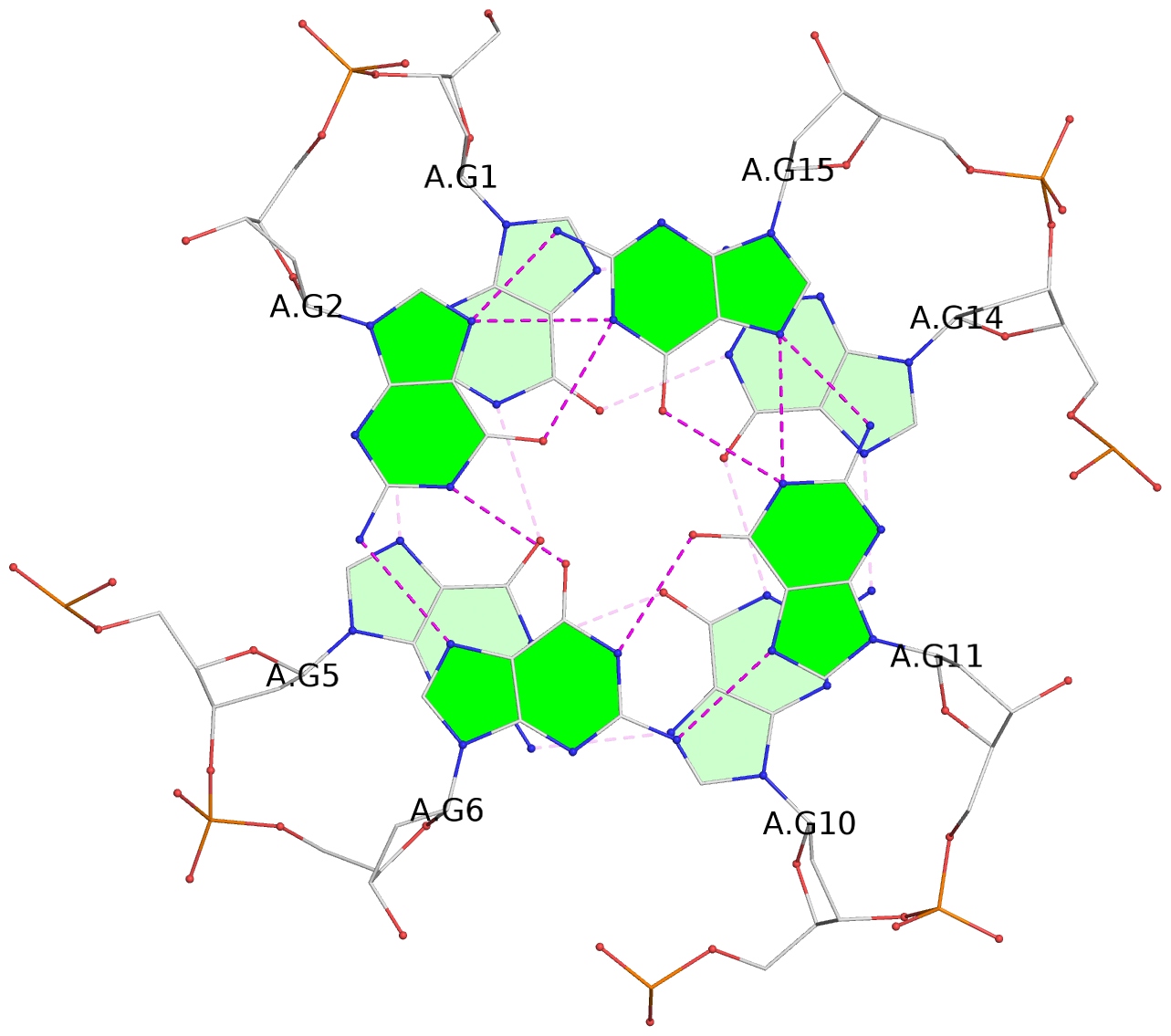
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.036 type=planar nts=4 GGGG A.DG1,A.DG5,A.DG10,A.DG14 2 glyco-bond=---- sugar=---- groove=---- planarity=0.063 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG11,A.DG15 3 glyco-bond=---- sugar=---- groove=---- planarity=0.037 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG12,A.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.