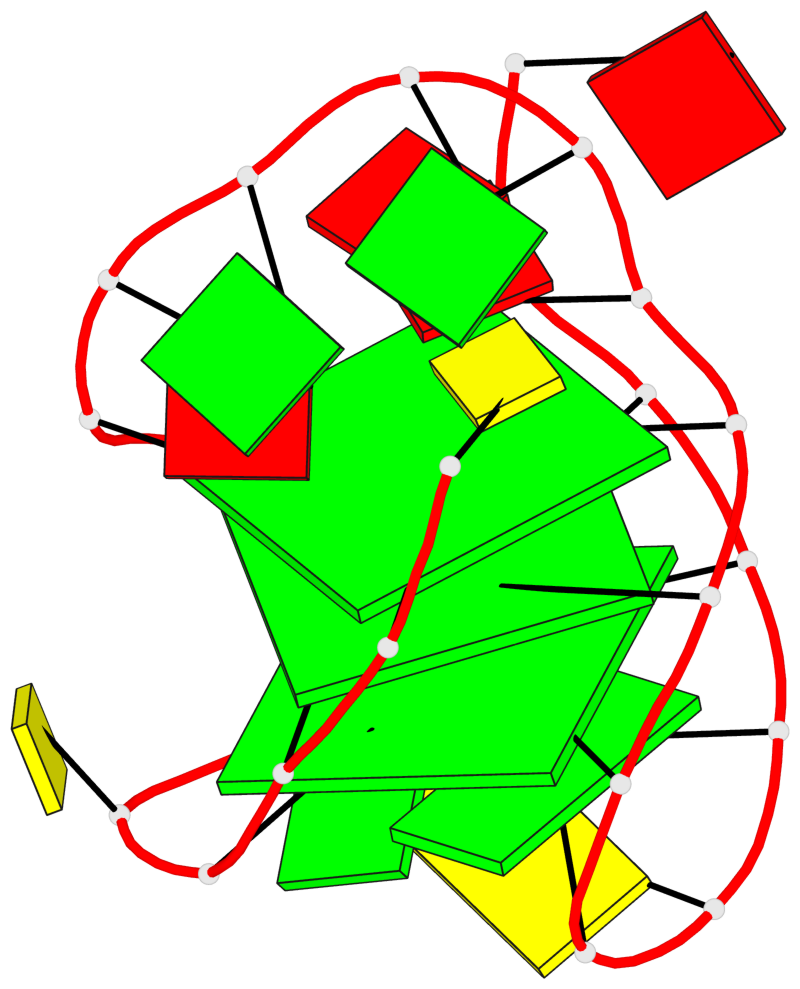
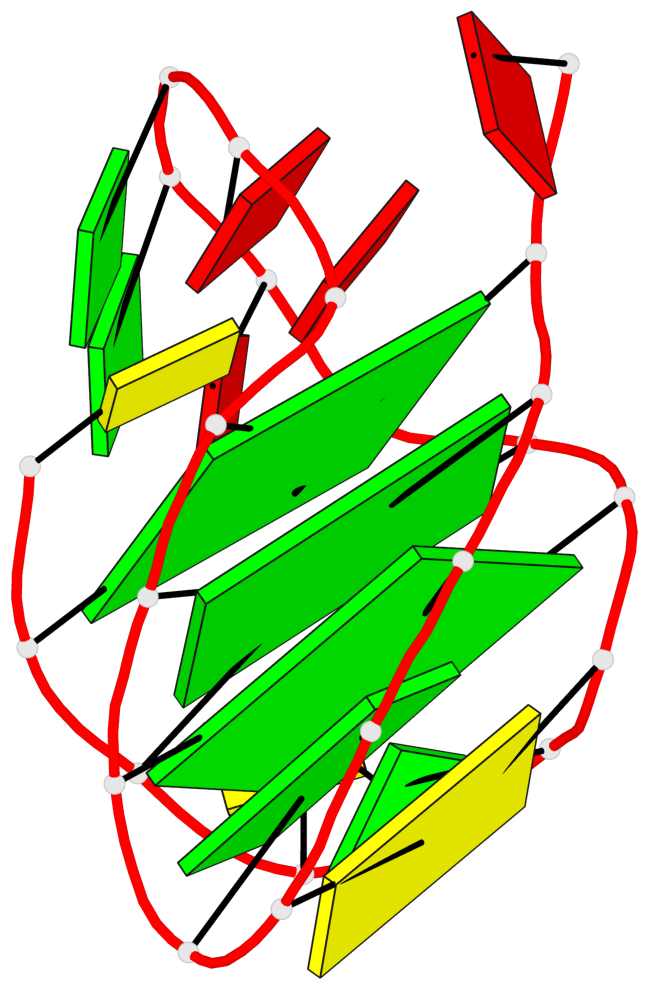
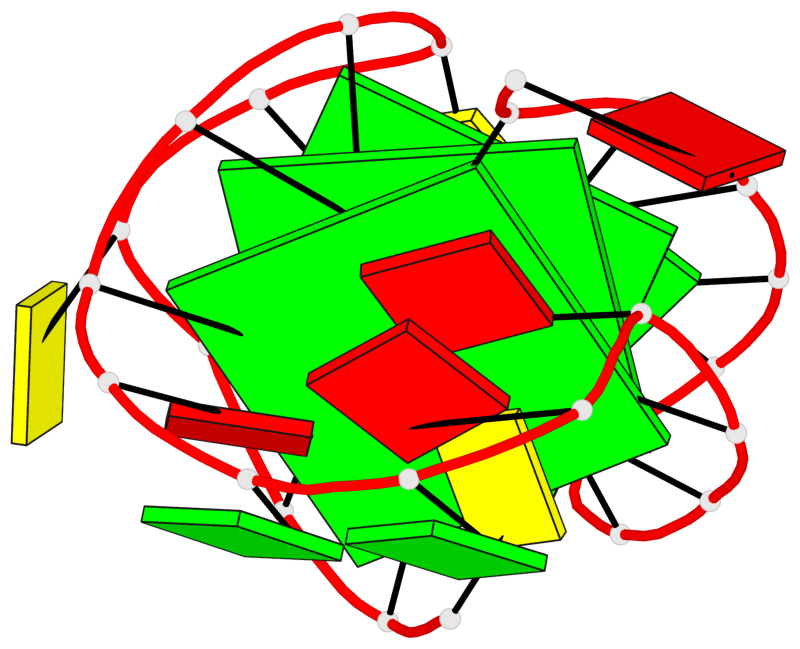
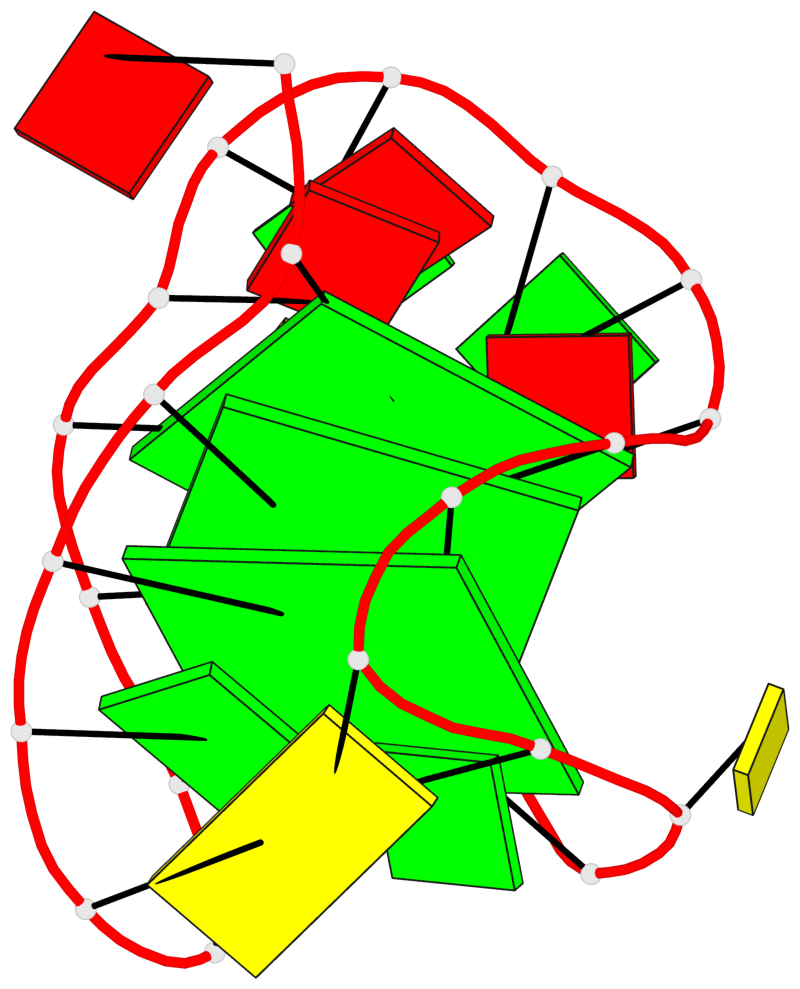
Detailed DSSR results for the G-quadruplex: PDB entry 6zx6
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6zx6
- Class
- DNA
- Method
- NMR
- Summary
- Antiparallel basket-type g-quadruplex DNA structure formed in human bcl-2 promoter containing 8-oxog
- Reference
- Bielskute S, Plavec J, Podbevsek P (2021): "Oxidative lesions modulate G-quadruplex stability and structure in the human BCL2 promoter." Nucleic Acids Res., 49, 2346-2356. doi: 10.1093/nar/gkab057.
- Abstract
- Misregulation of BCL2 expression has been observed with many diseases and is associated with cellular exposure to reactive oxygen species. A region upstream of the P1 promoter in the human BCL2 gene plays a major role in regulating transcription. This G/C-rich region is highly polymorphic and capable of forming G-quadruplex structures. Herein we report that an oxidative event simulated with an 8-oxo-7,8-dihydroguanine (oxoG) substitution within a long G-tract results in a reduction of structural polymorphism. Surprisingly, oxoG within a 25-nt construct boosts thermal stability of the resulting G-quadruplex. This is achieved by distinct hydrogen bonding properties of oxoG, which facilitate formation of an antiparallel basket-type G-quadruplex with a three G-quartet core and a G·oxoG·C base triad. While oxoG has previously been considered detrimental for G-quadruplex formation, its stabilizing effect within a promoter described in this study suggests a potential novel regulatory role of oxidative stress in general and specifically in BCL2 gene transcription.
- G4 notes
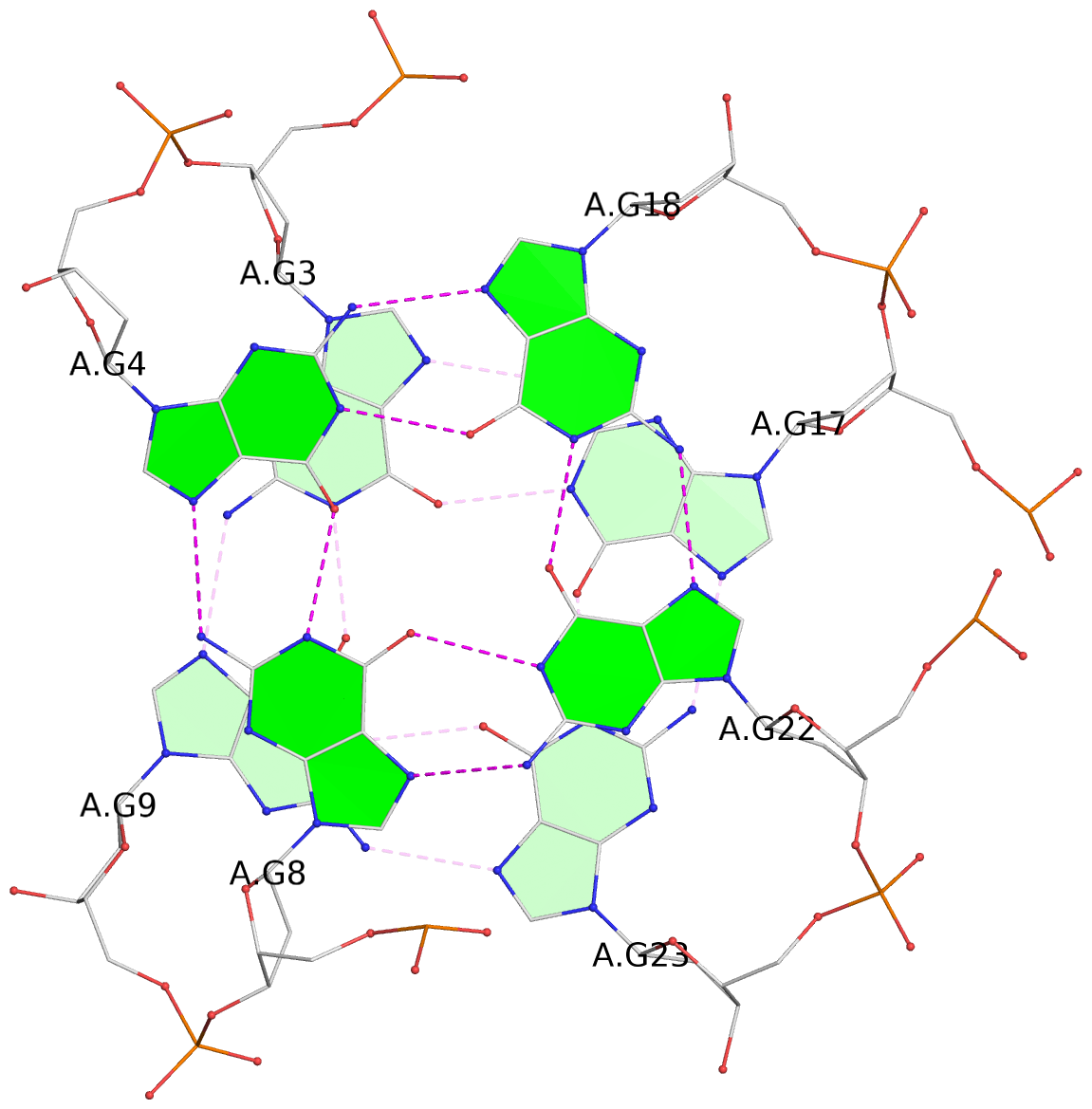
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-LwD+Ln), basket(2+2), UDDU
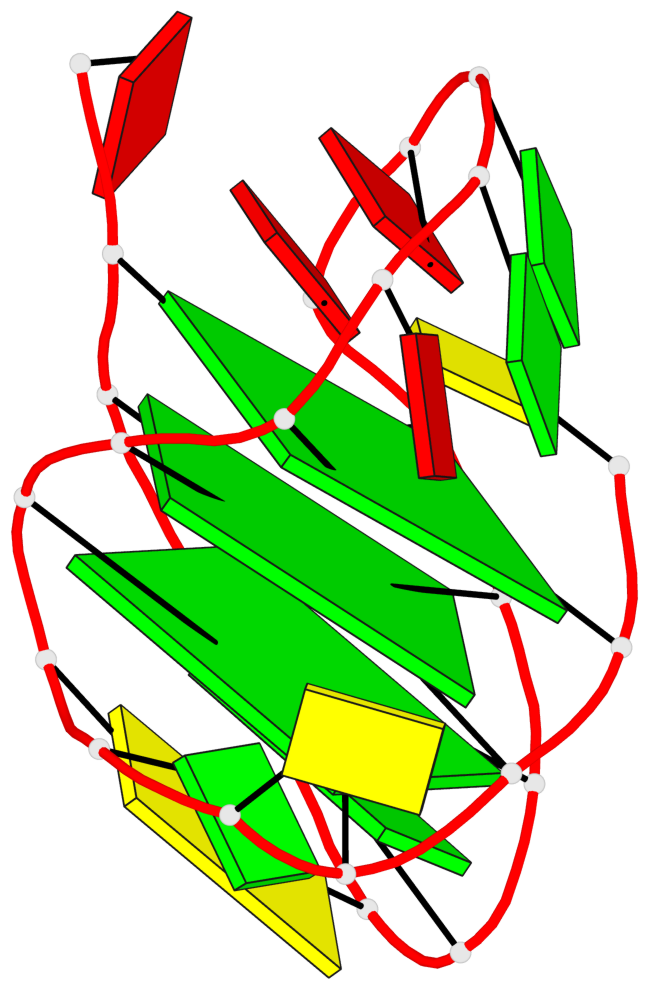
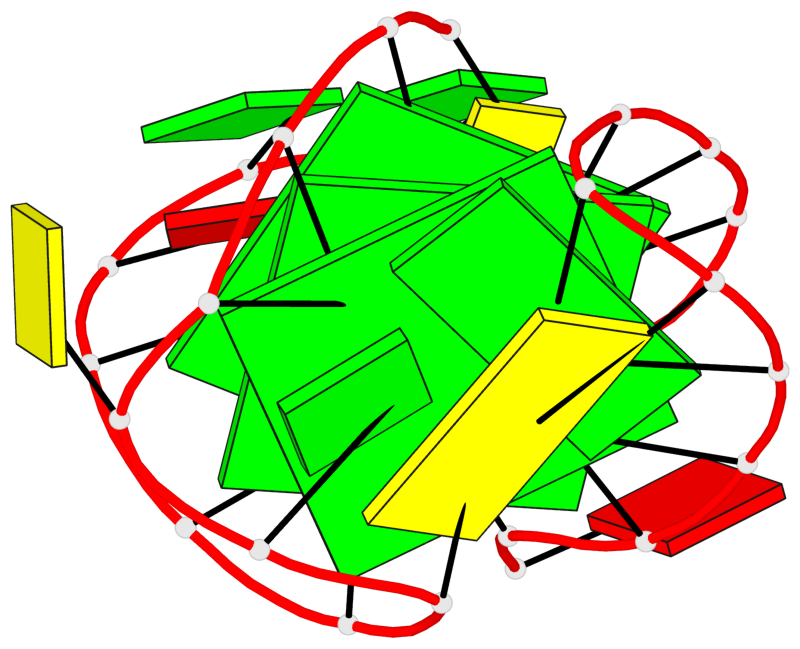
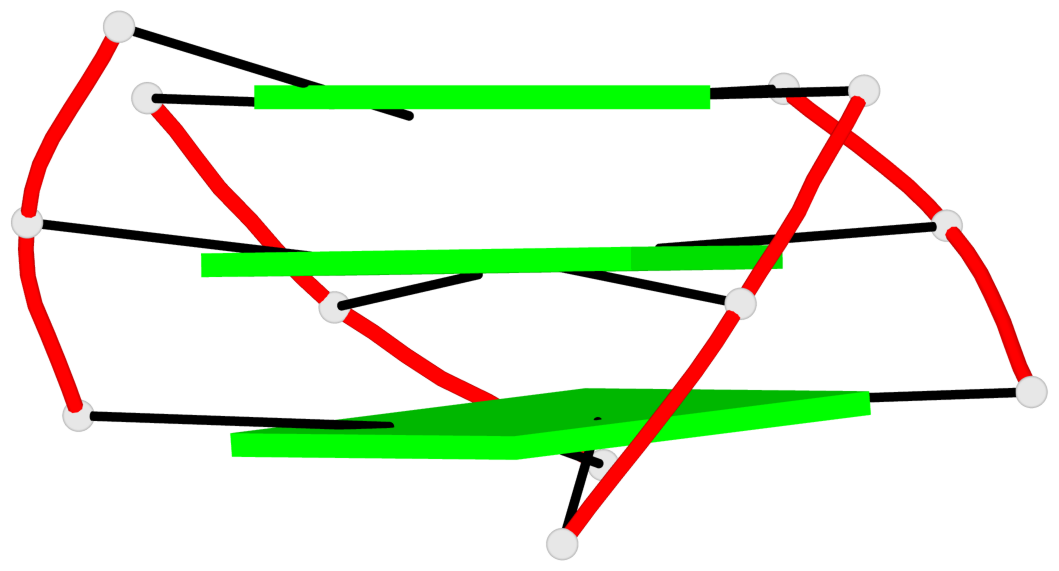
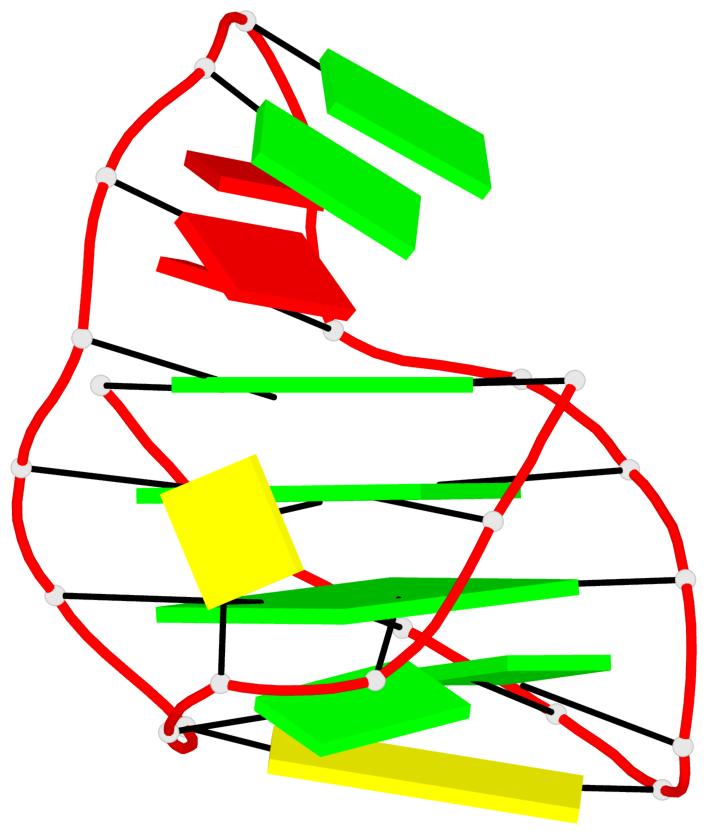
Base-block schematics in six views
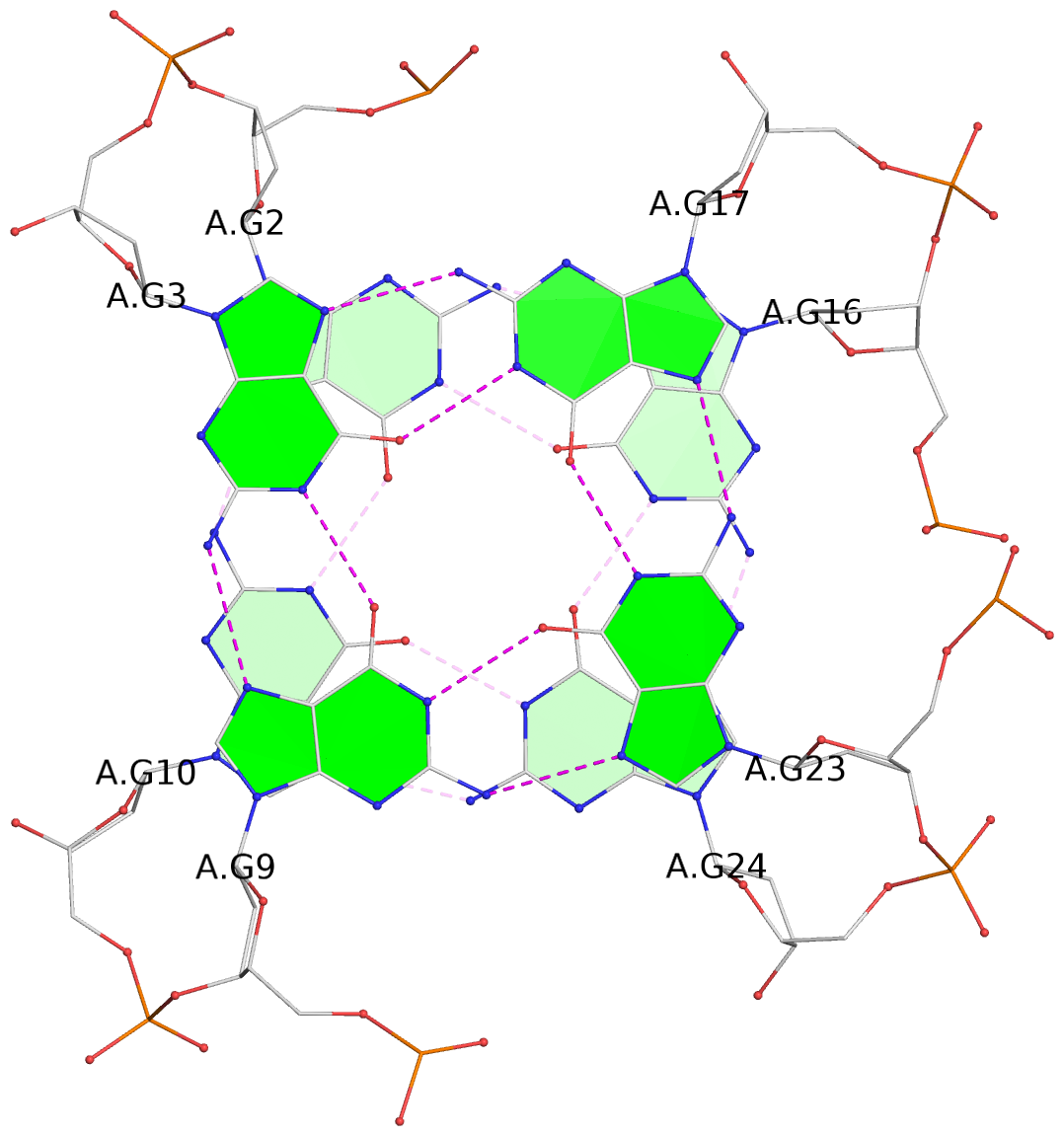
List of 3 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.394 type=other nts=4 GGGG A.DG2,A.DG10,A.DG24,A.DG16 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.423 type=other nts=4 GGGG A.DG3,A.DG9,A.DG23,A.DG17 3 glyco-bond=s--s sugar=-.-- groove=w-n- planarity=0.517 type=other nts=4 GGGG A.DG4,A.DG8,A.DG22,A.DG18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.