Detailed DSSR results for the G-quadruplex: PDB entry 7d33
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7d33
- Class
- DNA
- Method
- X-ray (2.117 Å)
- Summary
- The pb2+ complexed structure of tba g8c mutant
- Reference
- Liu H, Gao Y, Mathivanan J, Shen F, Chen X, Li Y, Shao Z, Zhang Y, Shao Q, Sheng J, Gan J (2022): "Structure-guided development of Pb 2+ -binding DNA aptamers." Sci Rep, 12, 460. doi: 10.1038/s41598-021-04243-2.
- Abstract
- Owing to its great threat to human health and environment, Pb2+ pollution has been recognized as a major public problem by the World Health Organization (WHO). Many DNA aptamers have been utilized in the development of Pb2+-detection sensors, but the underlying mechanisms remain elusive. Here, we report three Pb2+-complexed structures of the thrombin binding aptamer (TBA). These high-resolution crystal structures showed that TBA forms intramolecular G-quadruplex and Pb2+ is bound by the two G-tetrads in the center. Compared to K+-stabilized G-quadruplexes, the coordinating distance between Pb2+ and the G-tetrads are much shorter. The T3T4 and T12T13 linkers play important roles in dimerization and crystallization of TBA, but they are changeable for Pb2+-binding. In combination with mutagenesis and CD spectra, the G8C mutant structure unraveled that the T7G8T9 linker of TBA is also variable. In addition to expansion of the Pb2+-binding aptamer sequences, our study also set up one great example for quick and rational development of other aptamers with similar or optimized binding activity.
- G4 notes
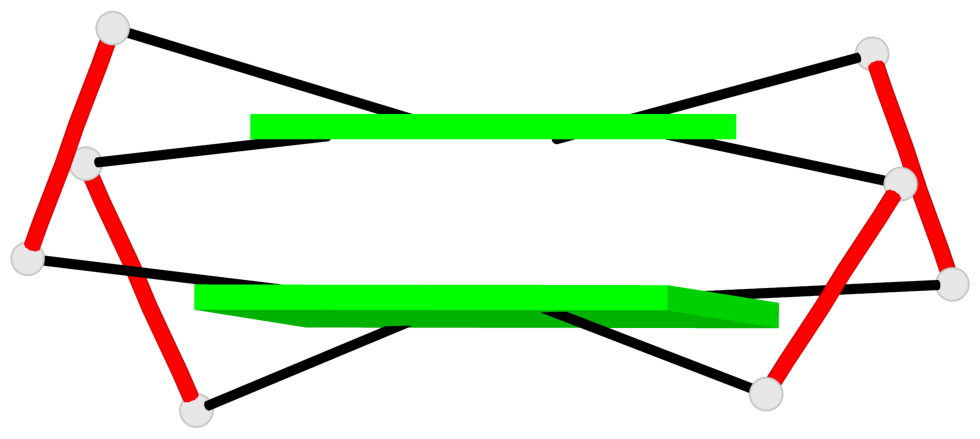
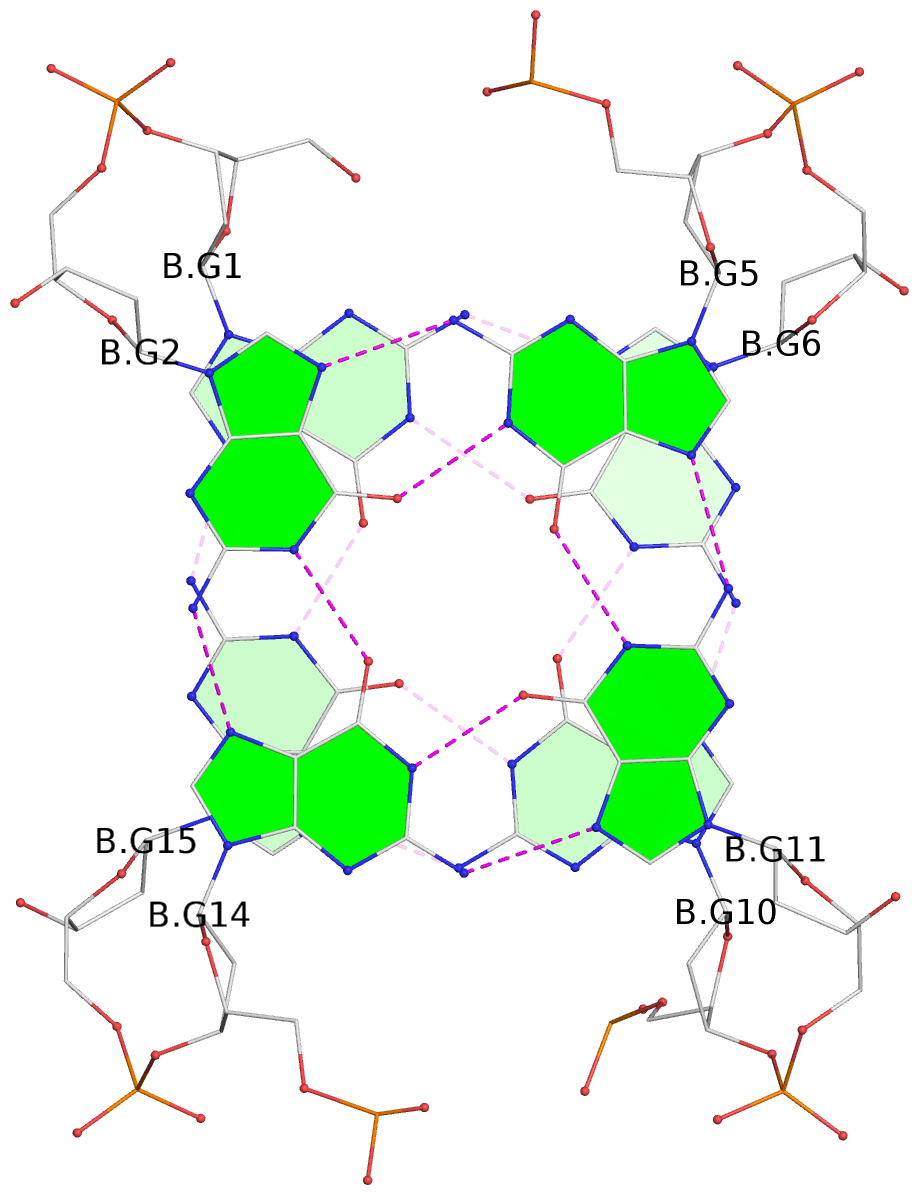
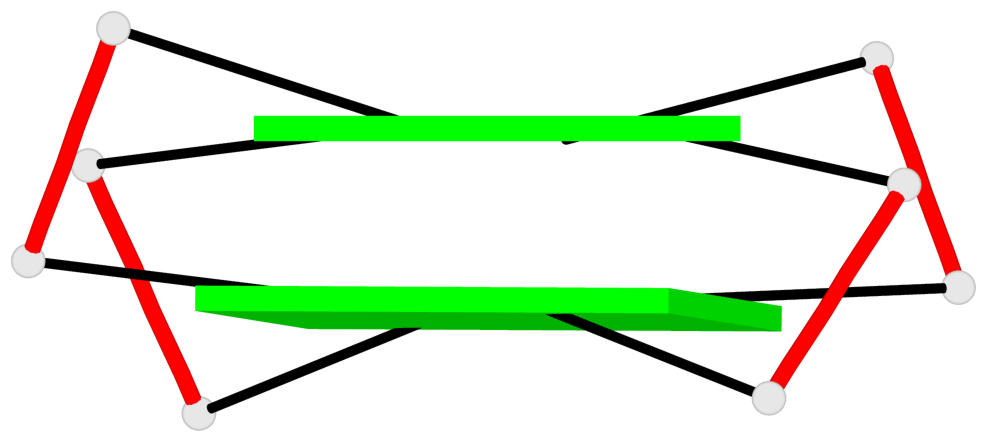
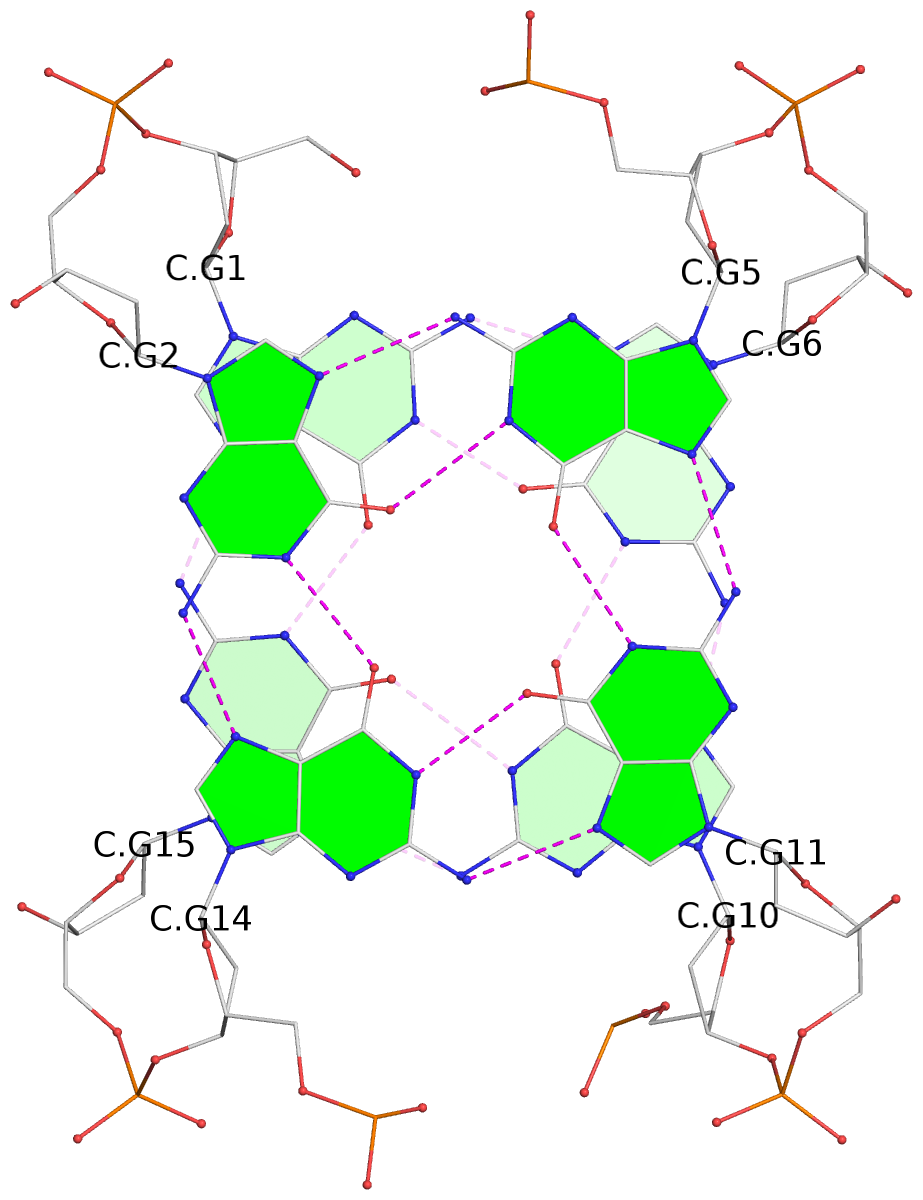
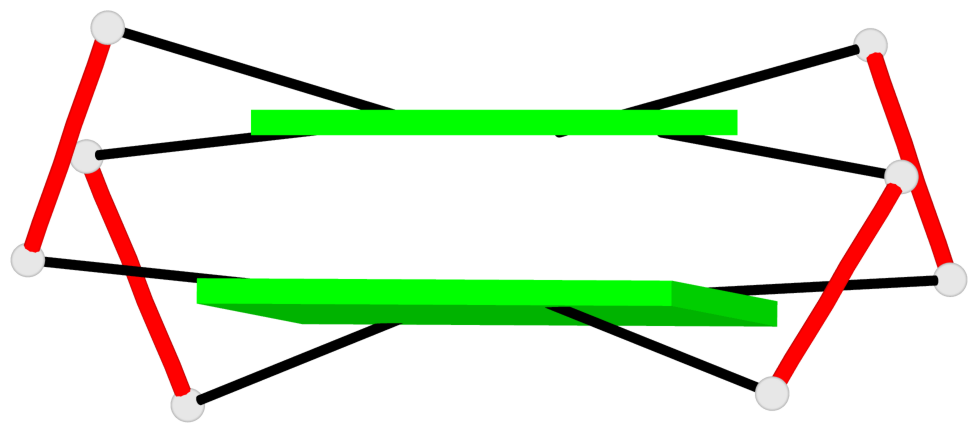
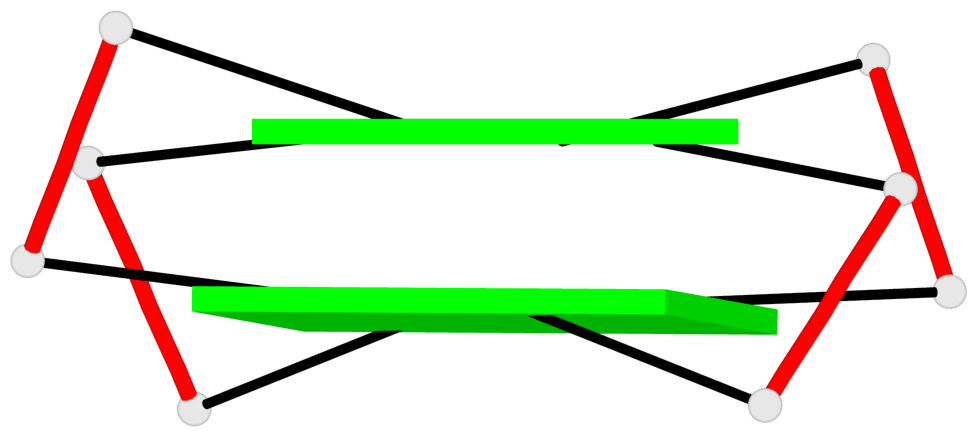
- 10 G-tetrads, 5 G4 helices, 5 G4 stems, 2(+Ln+Lw+Ln), chair(2+2), UDUD
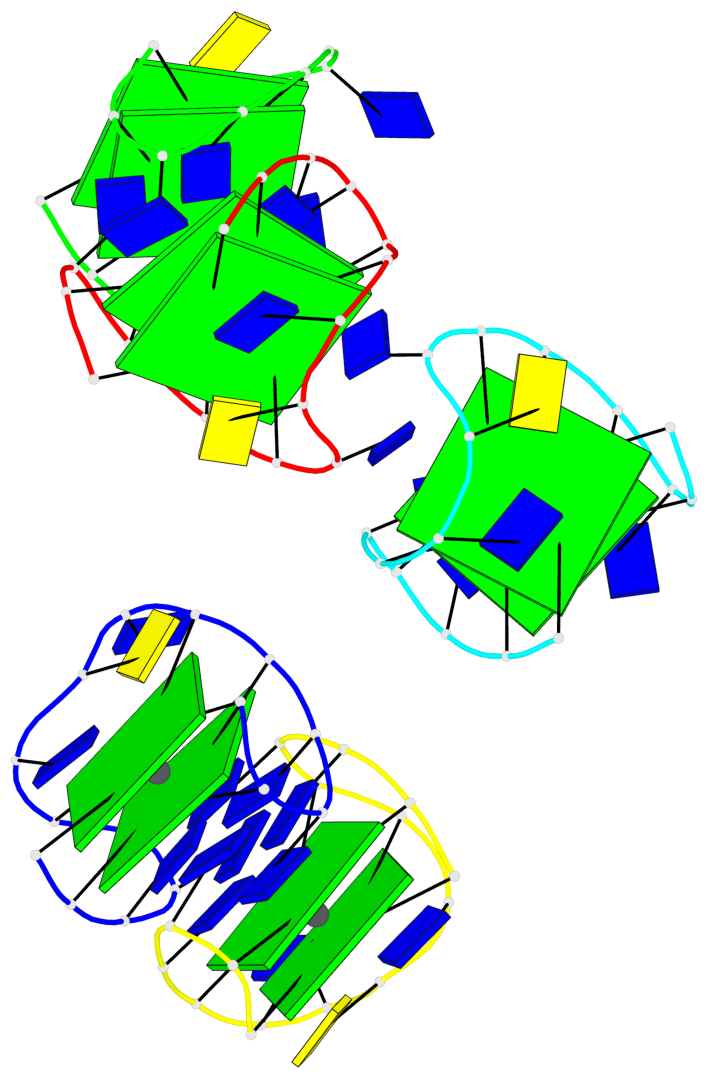
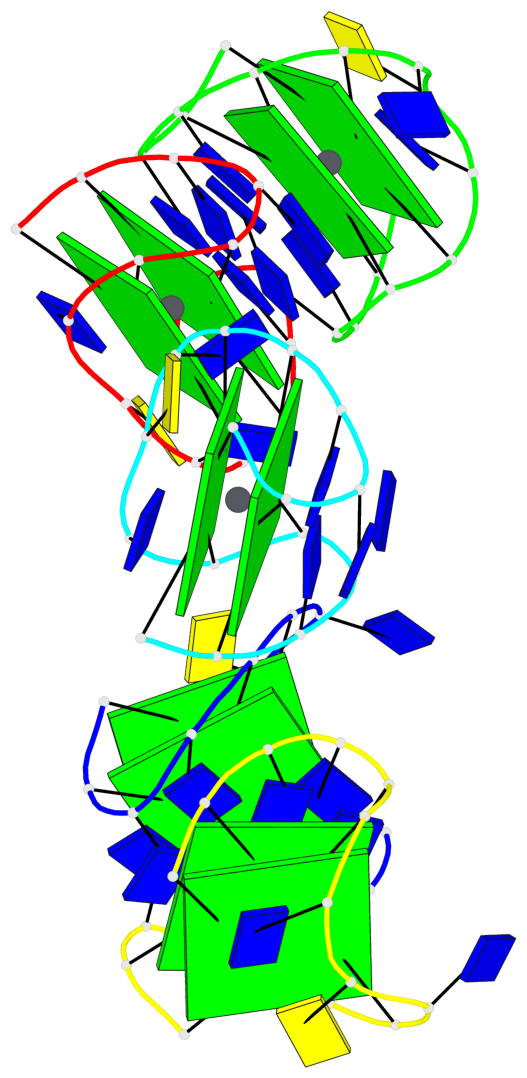
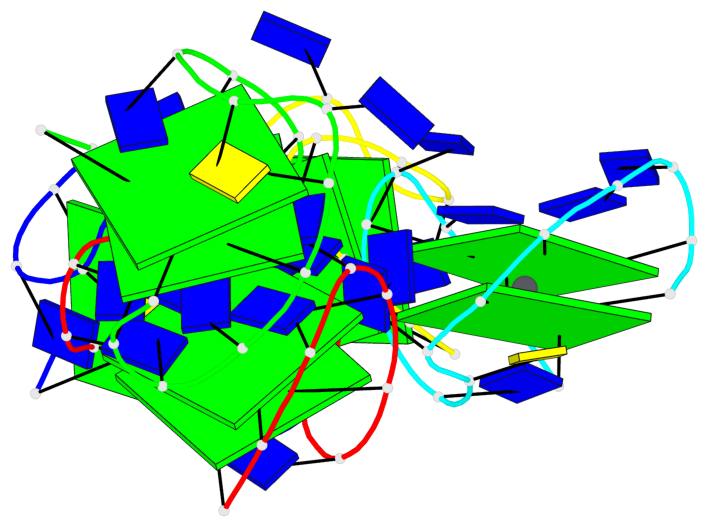
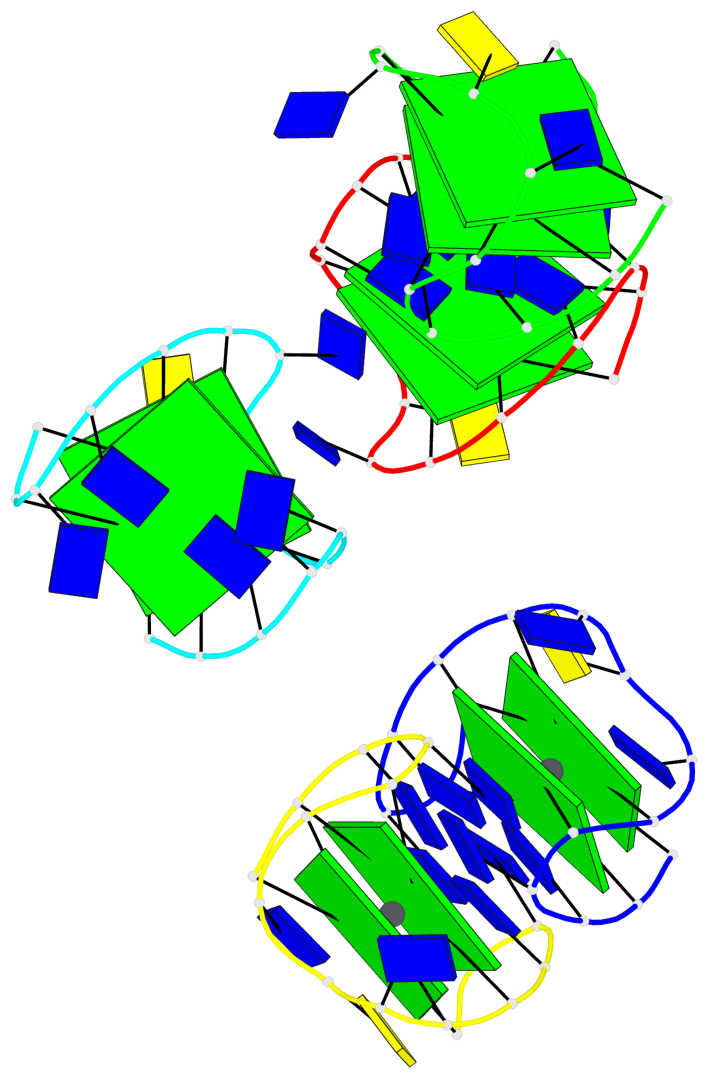
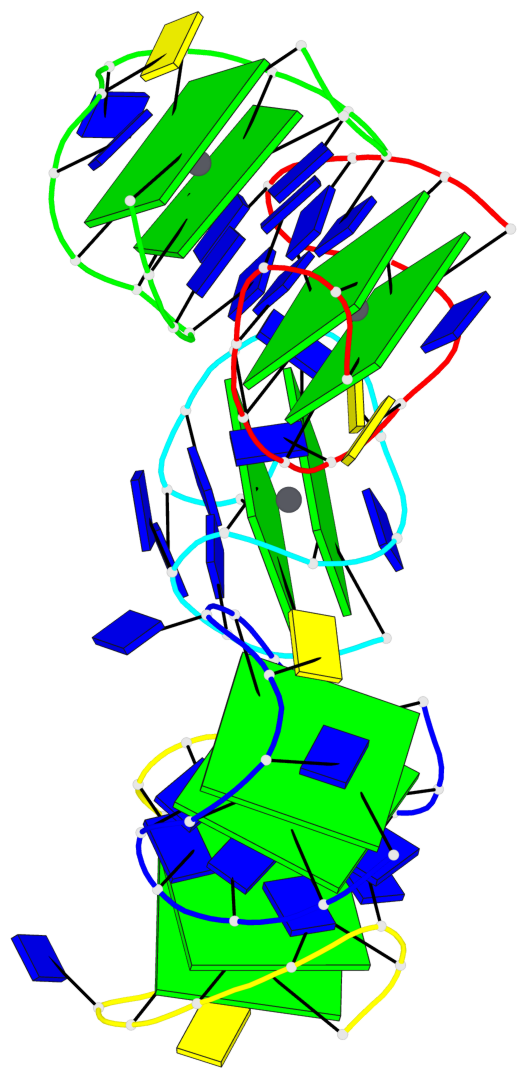
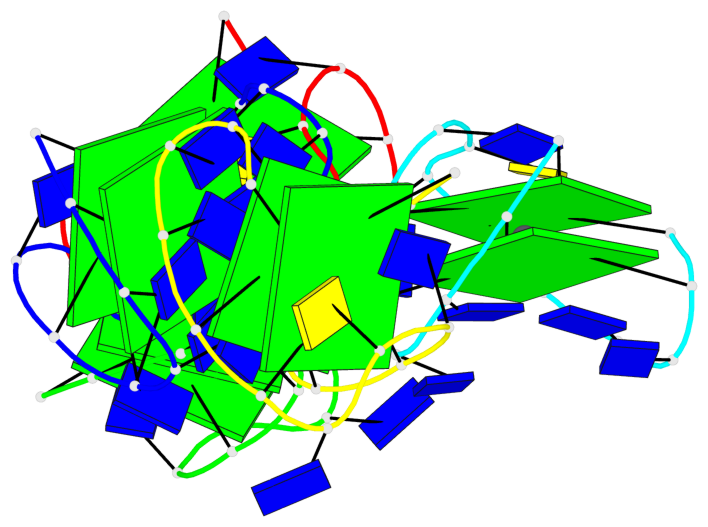
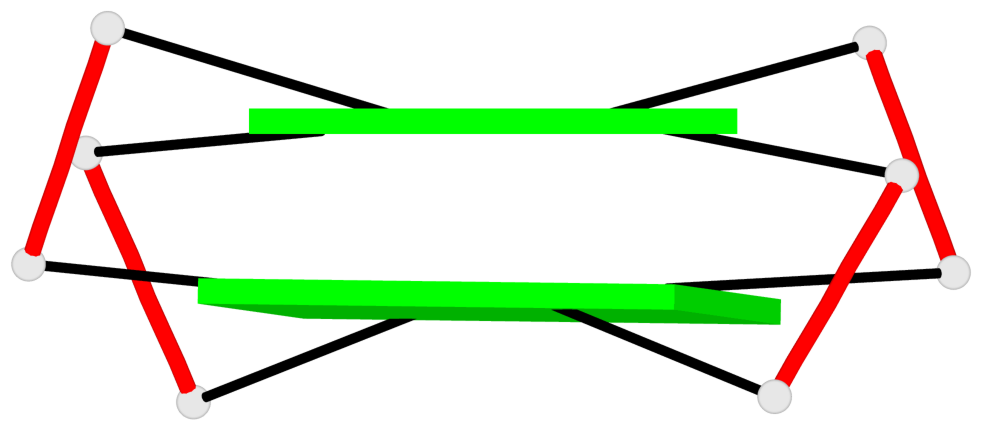
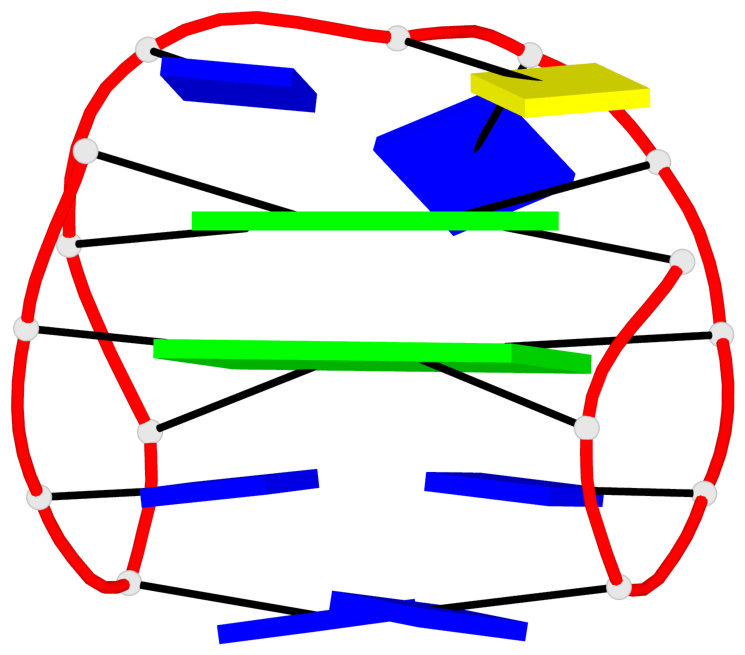
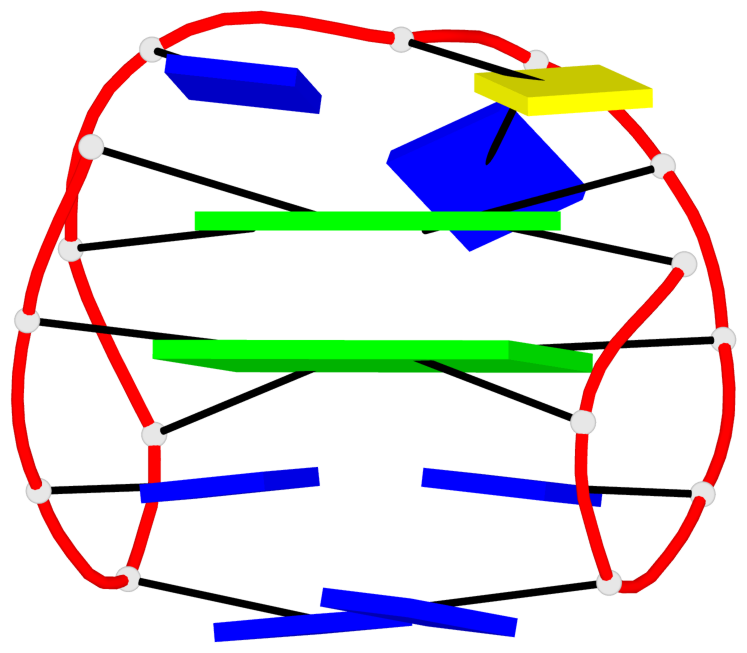
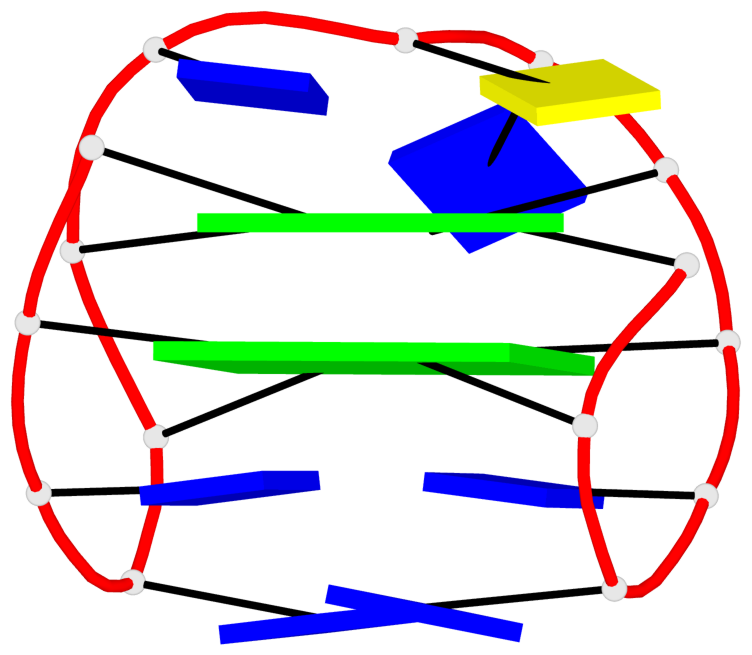
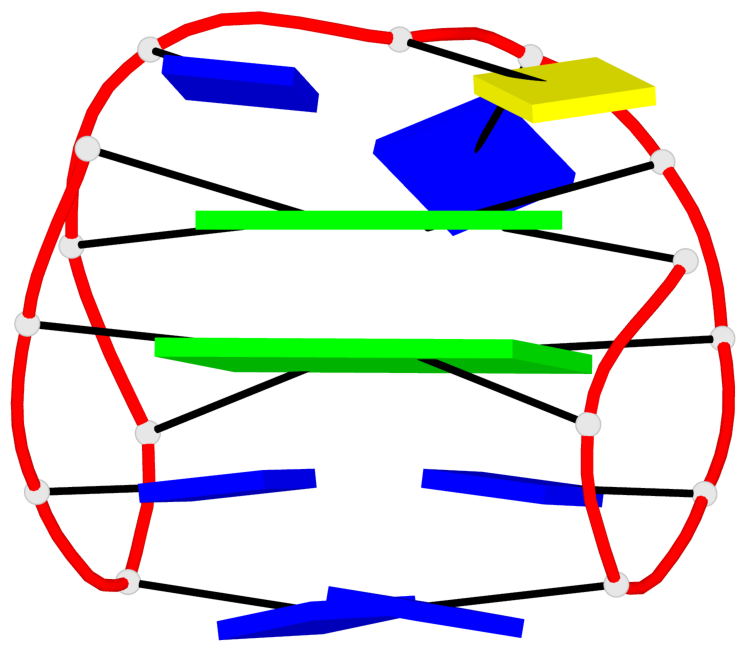
Base-block schematics in six views
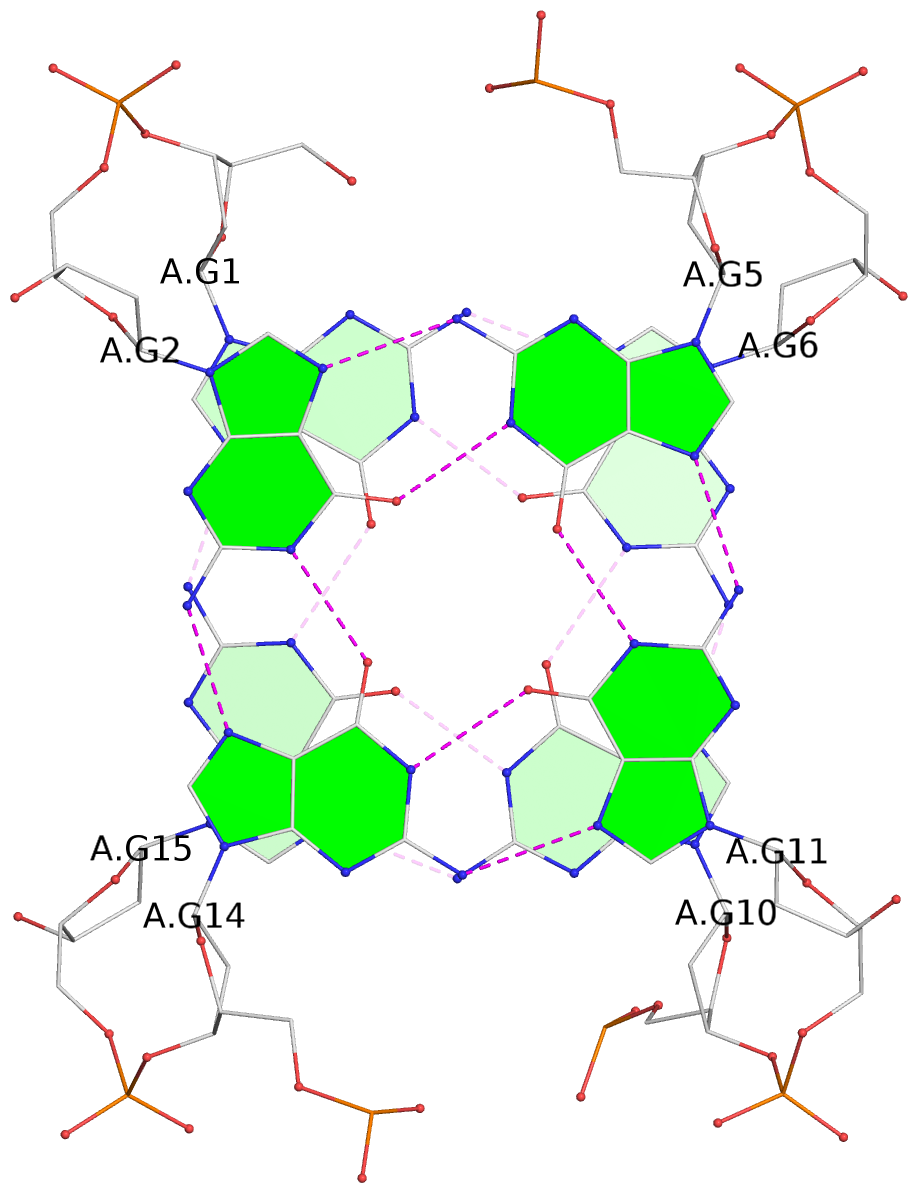
List of 10 G-tetrads
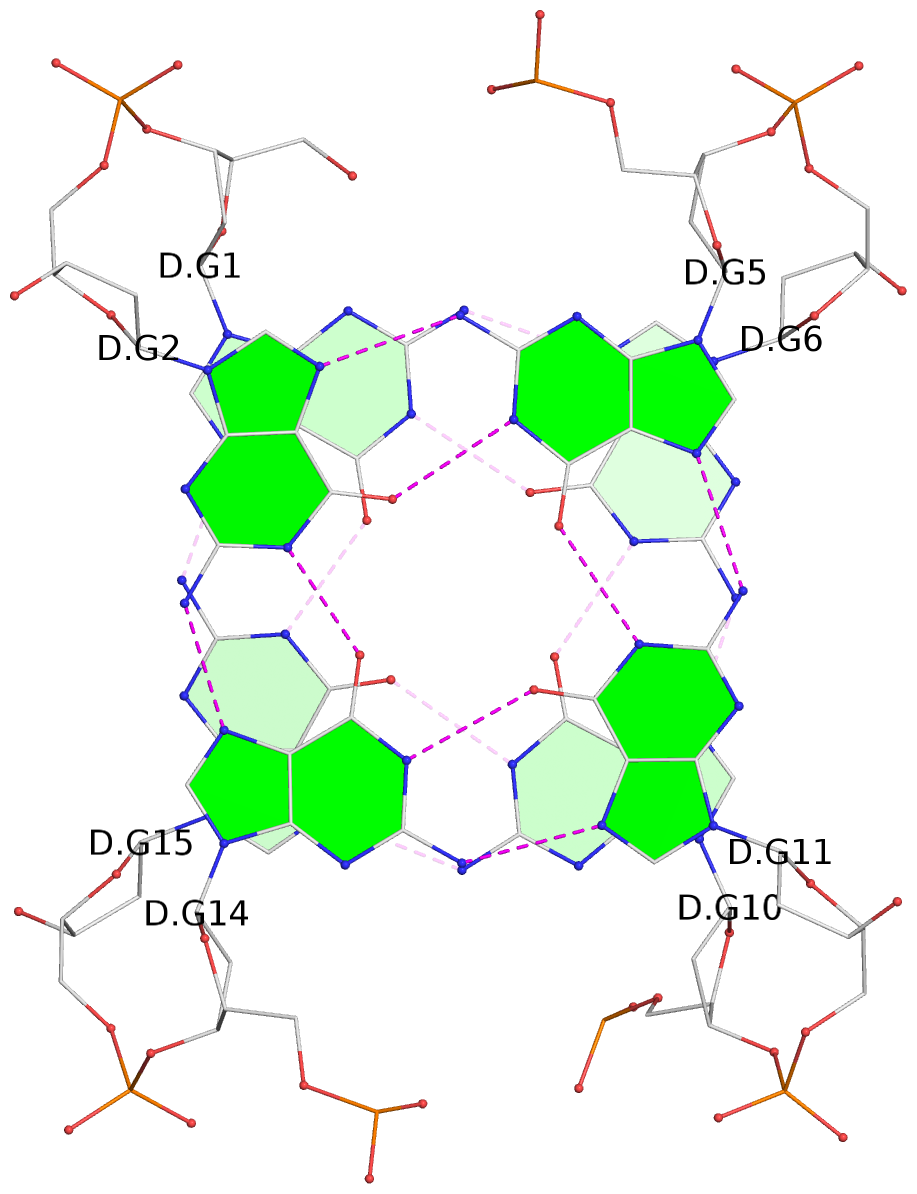
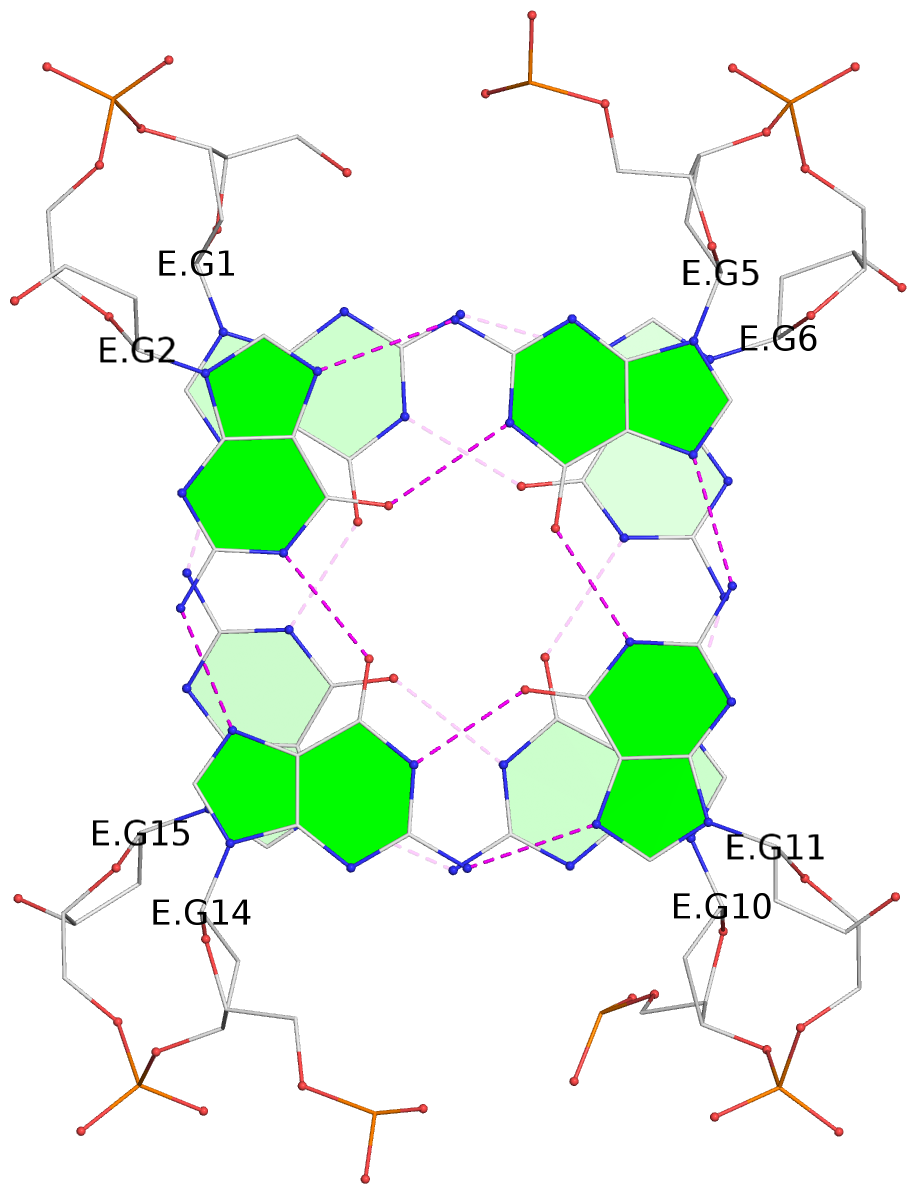
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.397 type=bowl-2 nts=4 GGGG A.DG1,A.DG15,A.DG10,A.DG6 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.442 type=bowl-2 nts=4 GGGG A.DG2,A.DG14,A.DG11,A.DG5 3 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.428 type=bowl-2 nts=4 GGGG B.DG1,B.DG15,B.DG10,B.DG6 4 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.431 type=bowl-2 nts=4 GGGG B.DG2,B.DG14,B.DG11,B.DG5 5 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.403 type=bowl-2 nts=4 GGGG C.DG1,C.DG15,C.DG10,C.DG6 6 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.432 type=bowl-2 nts=4 GGGG C.DG2,C.DG14,C.DG11,C.DG5 7 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.406 type=bowl-2 nts=4 GGGG D.DG1,D.DG15,D.DG10,D.DG6 8 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.420 type=bowl-2 nts=4 GGGG D.DG2,D.DG14,D.DG11,D.DG5 9 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.412 type=bowl-2 nts=4 GGGG E.DG1,E.DG15,E.DG10,E.DG6 10 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.419 type=bowl-2 nts=4 GGGG E.DG2,E.DG14,E.DG11,E.DG5
List of 5 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 2 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#3, 2 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#4, 2 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#5, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 5 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.