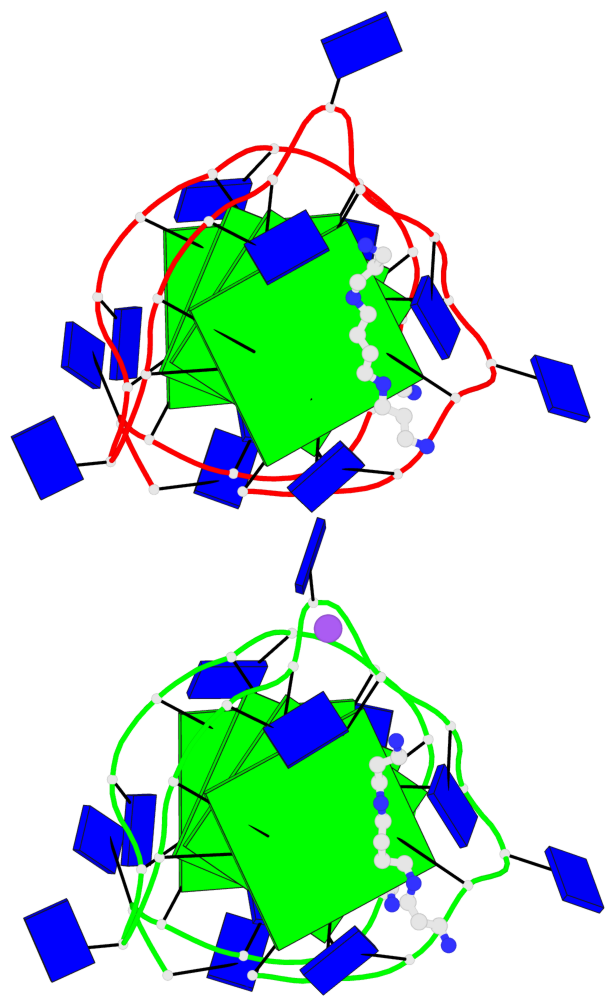
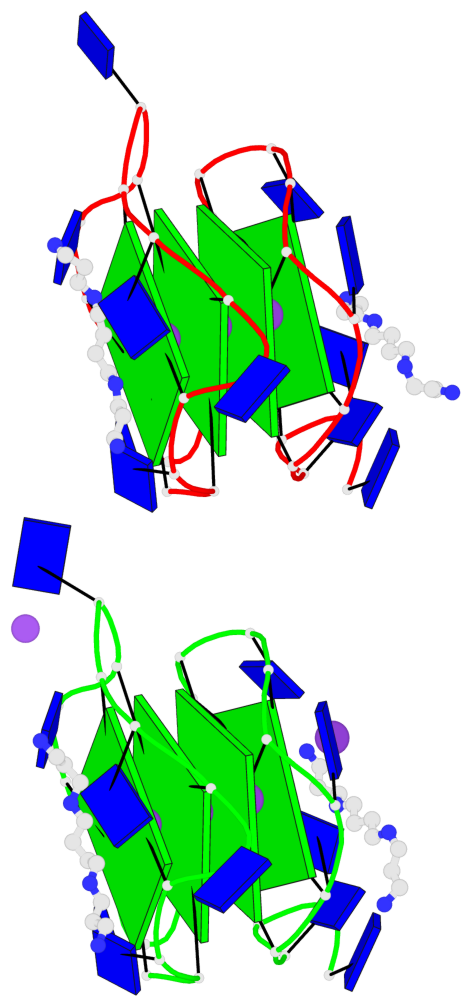
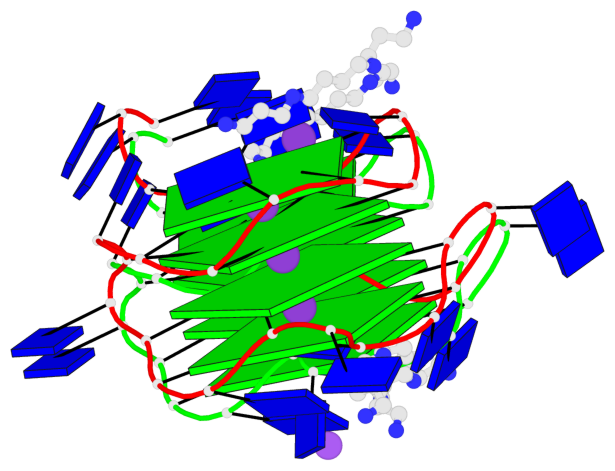
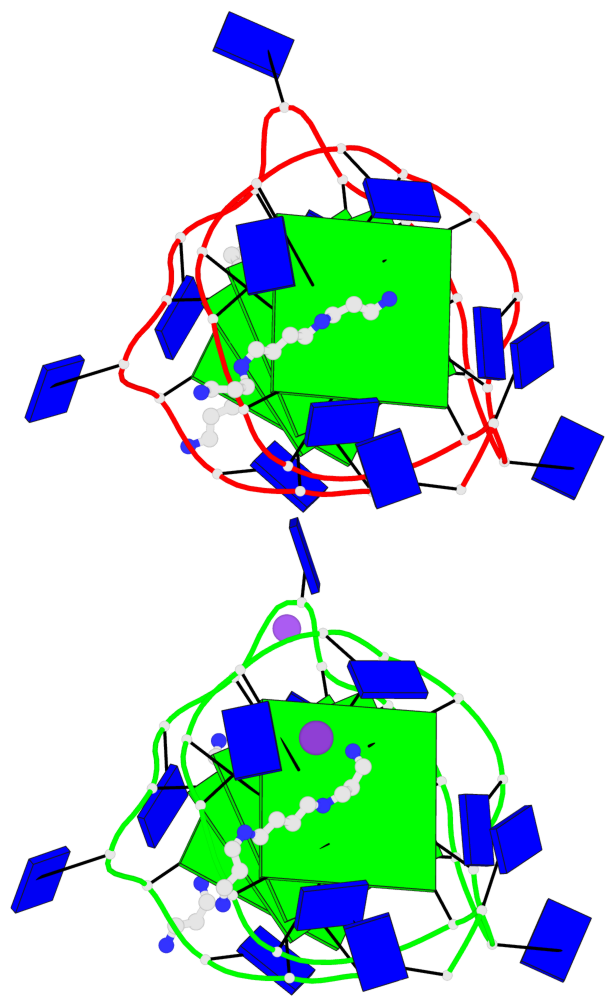
Detailed DSSR results for the G-quadruplex: PDB entry 7d5e
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7d5e
- Class
- DNA
- Method
- X-ray (1.296 Å)
- Summary
- Left-handed g-quadruplex containing two bulges
- Reference
- Das P, Ngo KH, Winnerdy FR, Maity A, Bakalar B, Mechulam Y, Schmitt E, Phan AT (2021): "Bulges in left-handed G-quadruplexes." Nucleic Acids Res., 49, 1724-1736. doi: 10.1093/nar/gkaa1259.
- Abstract
- G-quadruplex (G4) DNA structures with a left-handed backbone progression have unique and conserved structural features. Studies on sequence dependency of the structures revealed the prerequisites and some minimal motifs required for left-handed G4 formation. To extend the boundaries, we explore the adaptability of left-handed G4s towards the existence of bulges. Here we present two X-ray crystal structures and an NMR solution structure of left-handed G4s accommodating one, two and three bulges. Bulges in left-handed G4s show distinct characteristics as compared to those in right-handed G4s. The elucidation of intricate structural details will help in understanding the possible roles and limitations of these unique structures.
- G4 notes
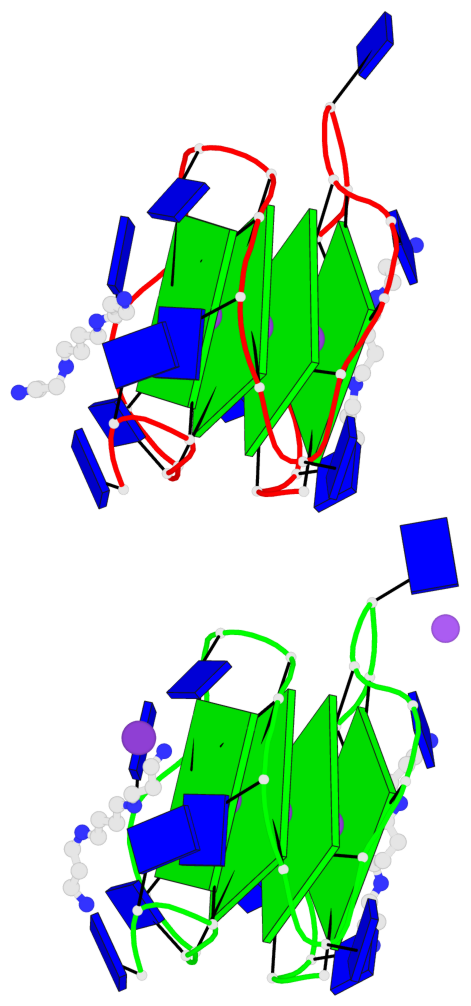
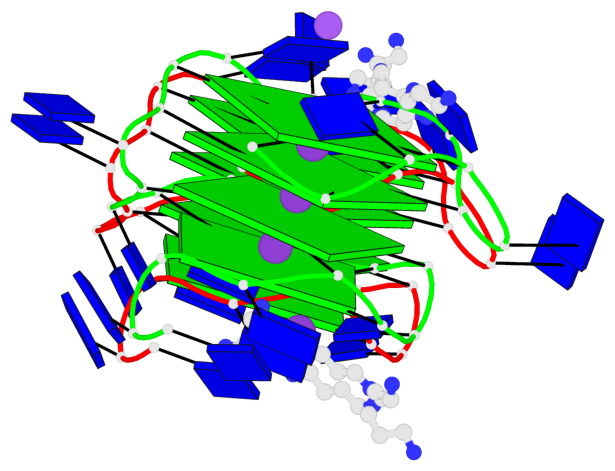
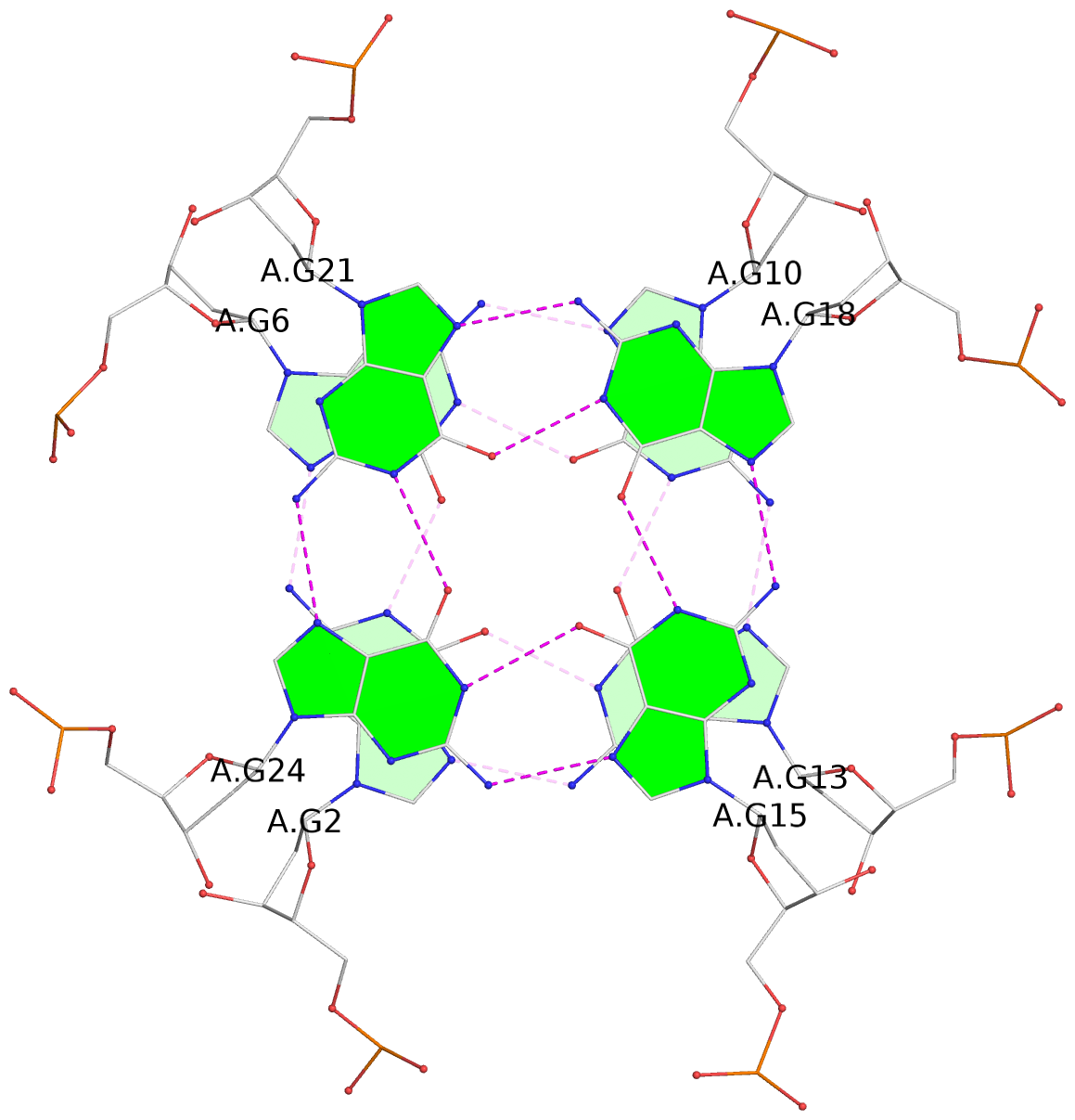
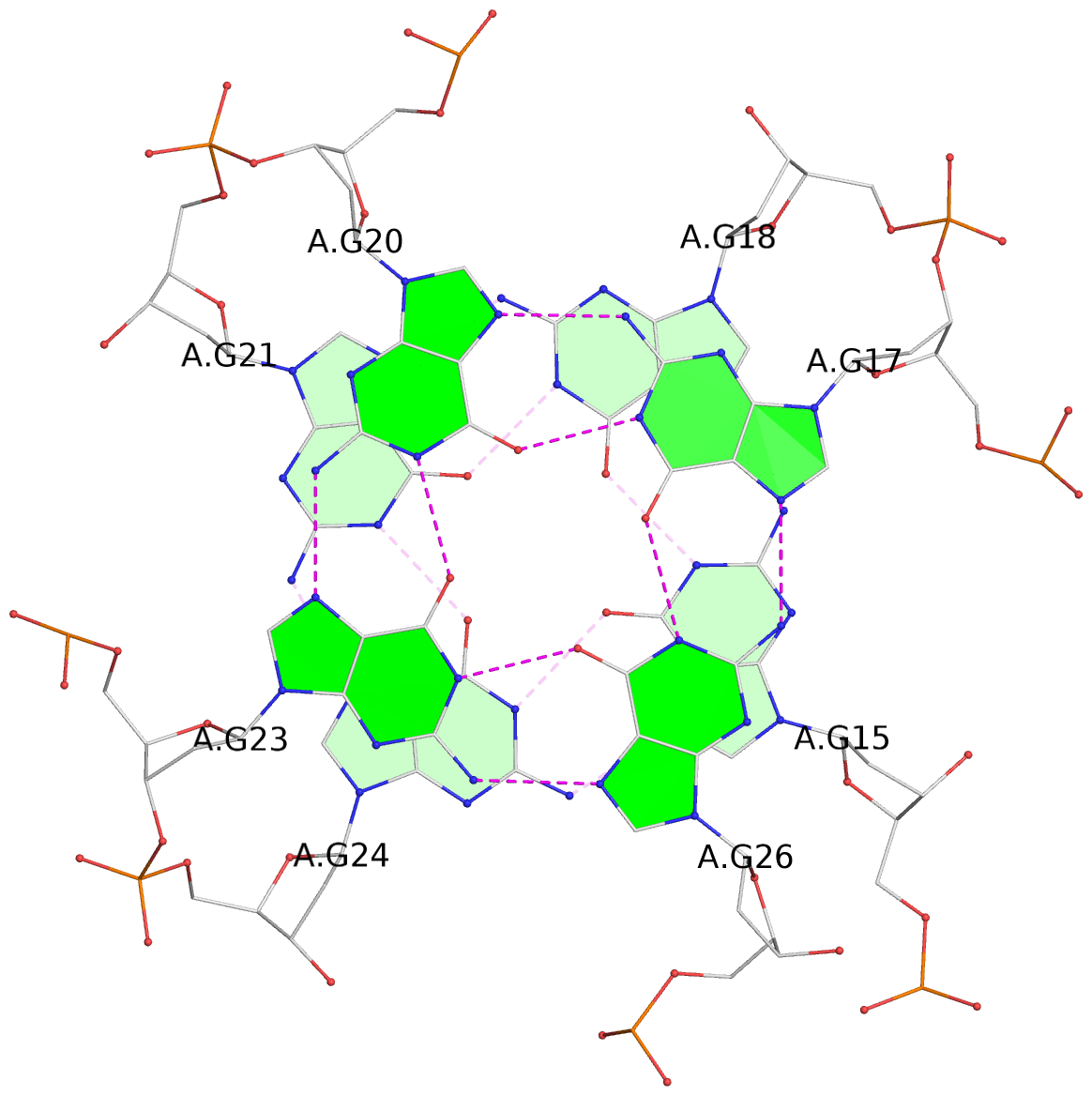
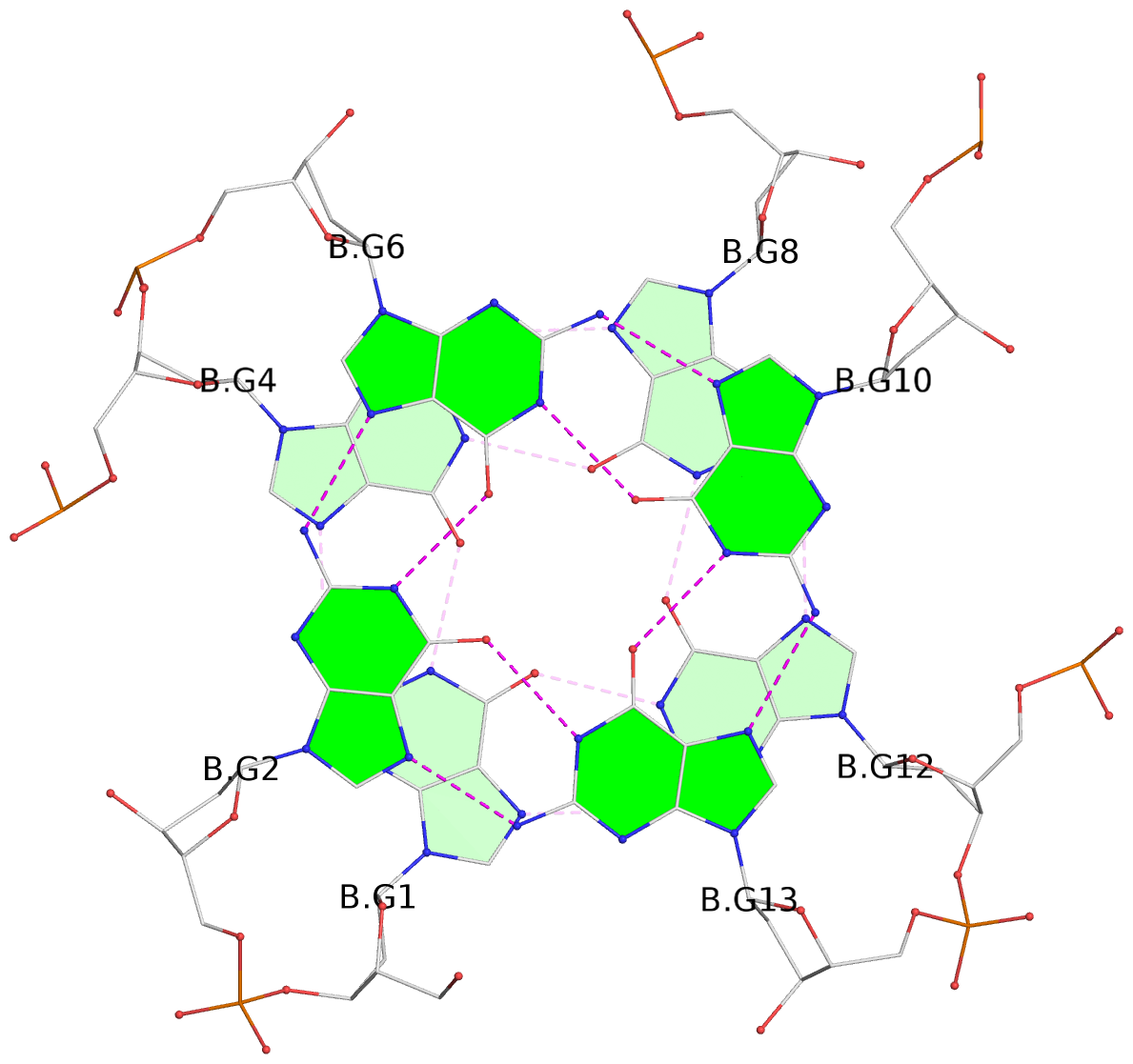
- 8 G-tetrads, 2 G4 helices, 2 G4 stems, 2(+P+P+P), parallel(4+0), UUUU, negative twist
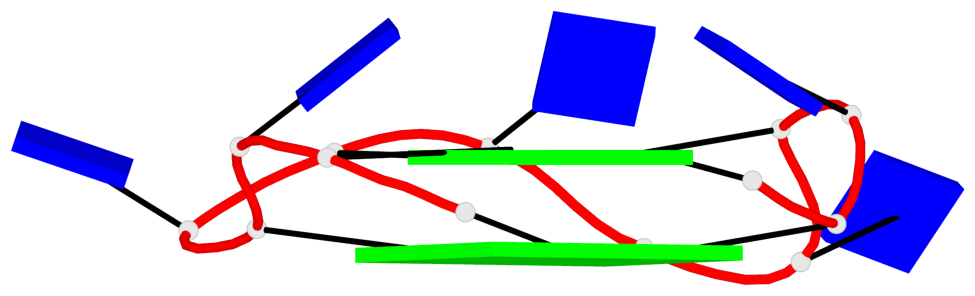
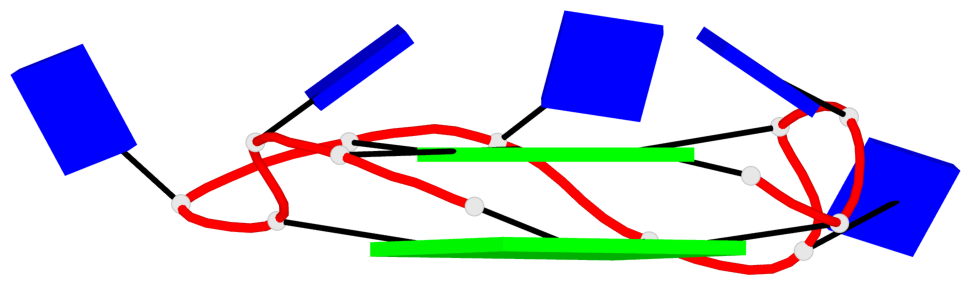
Base-block schematics in six views
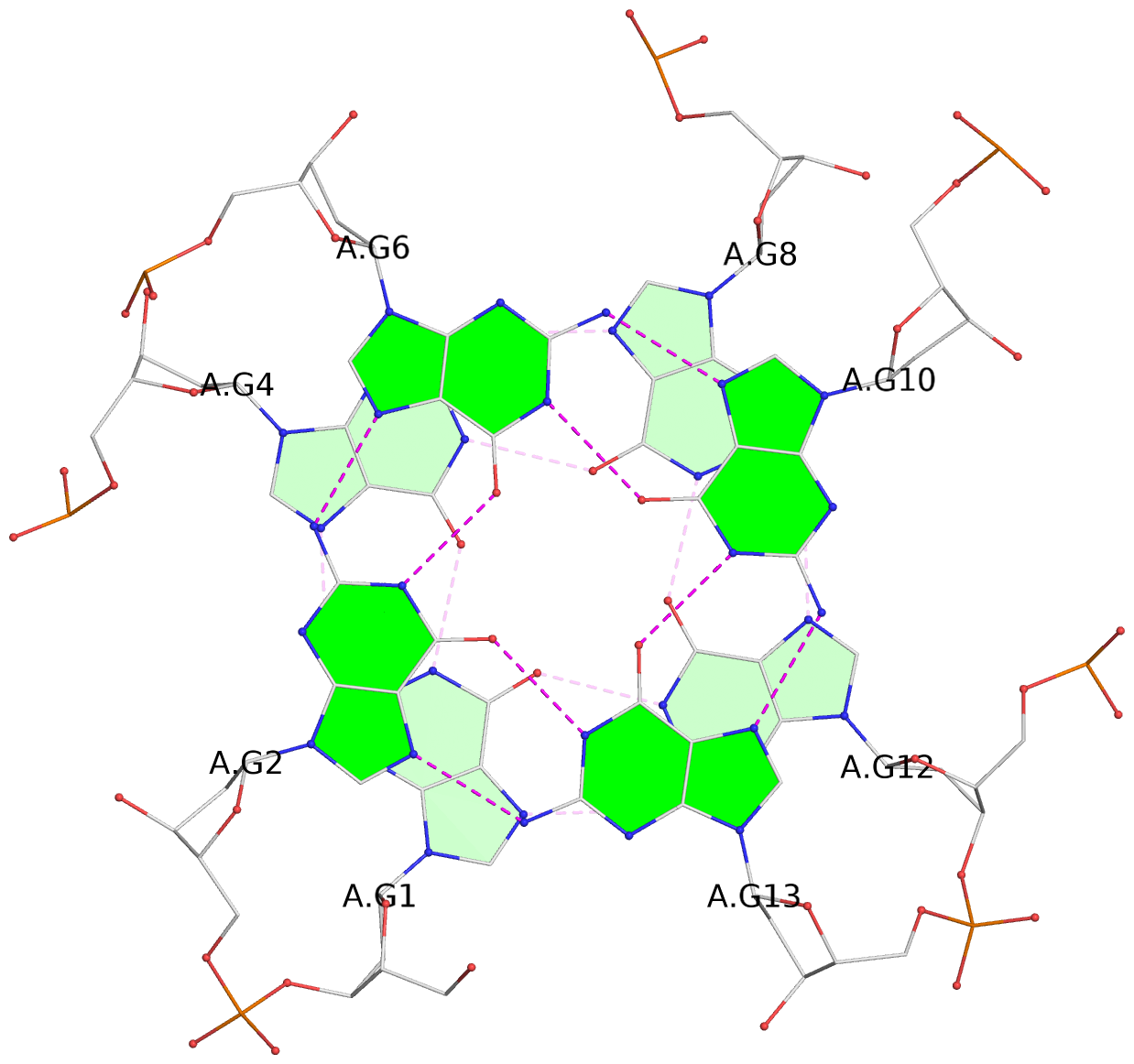
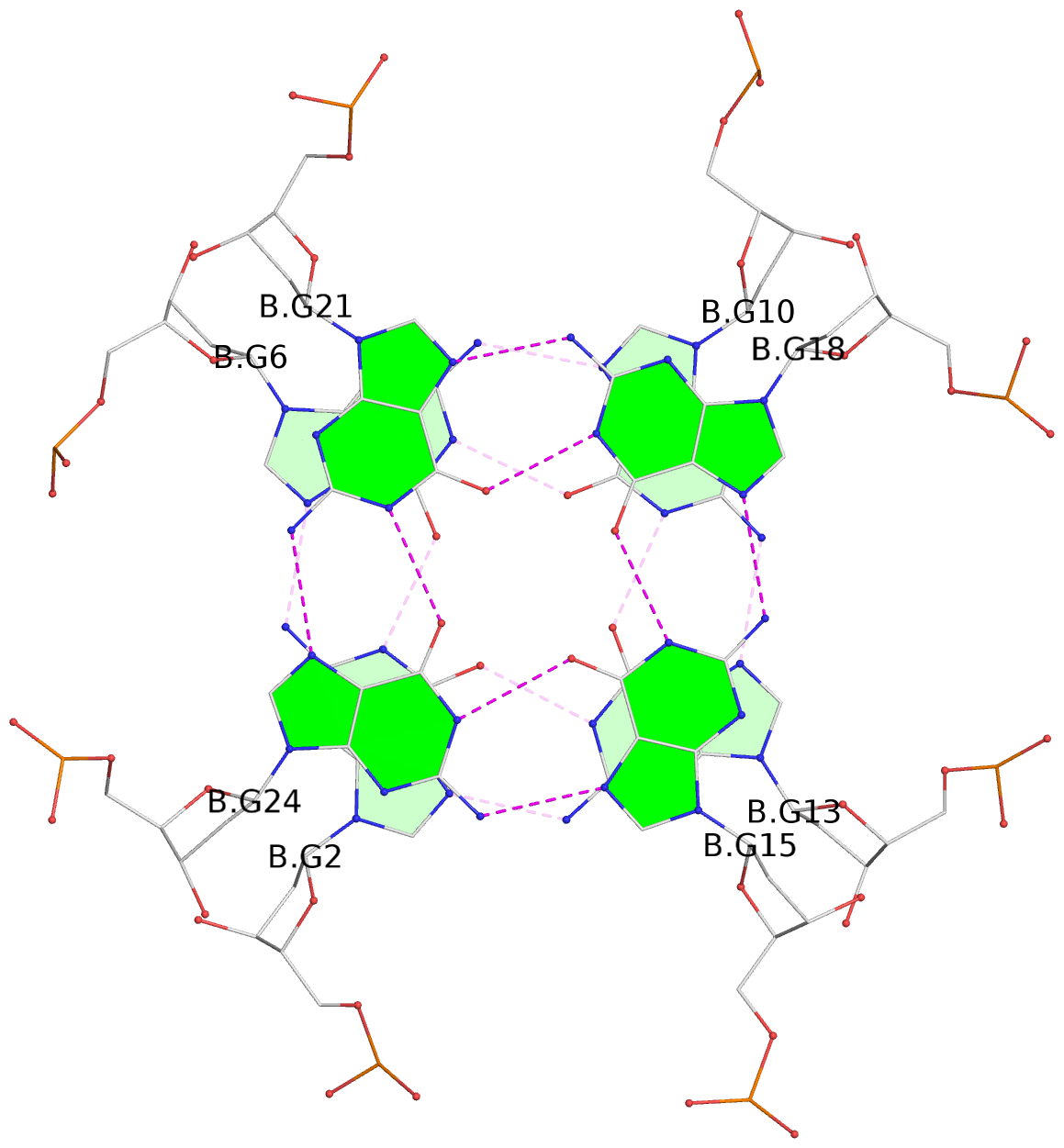
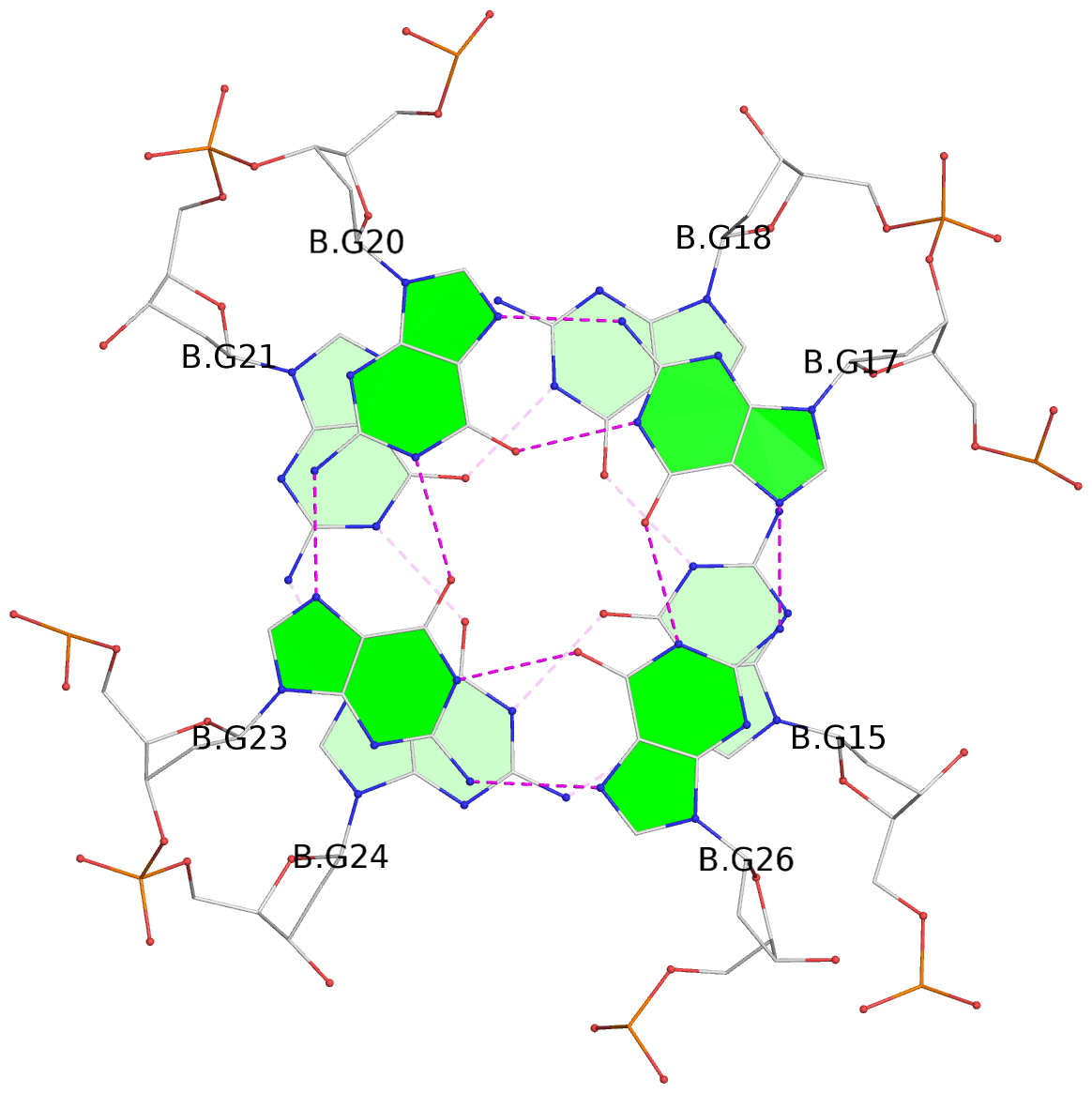
List of 8 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.137 type=planar nts=4 GGGG A.DG1,A.DG4,A.DG8,A.DG12 2 glyco-bond=---- sugar=---- groove=---- planarity=0.027 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG10,A.DG13 3 glyco-bond=---- sugar=---- groove=---- planarity=0.079 type=planar nts=4 GGGG A.DG15,A.DG18,A.DG21,A.DG24 4 glyco-bond=---- sugar=---- groove=---- planarity=0.197 type=other nts=4 GGGG A.DG17,A.DG20,A.DG23,A.DG26 5 glyco-bond=---- sugar=---- groove=---- planarity=0.124 type=planar nts=4 GGGG B.DG1,B.DG4,B.DG8,B.DG12 6 glyco-bond=---- sugar=---- groove=---- planarity=0.052 type=planar nts=4 GGGG B.DG2,B.DG6,B.DG10,B.DG13 7 glyco-bond=---- sugar=---- groove=---- planarity=0.095 type=planar nts=4 GGGG B.DG15,B.DG18,B.DG21,B.DG24 8 glyco-bond=---- sugar=---- groove=---- planarity=0.188 type=other nts=4 GGGG B.DG17,B.DG20,B.DG23,B.DG26
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(+P+P+P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(+P+P+P), parallel(4+0)
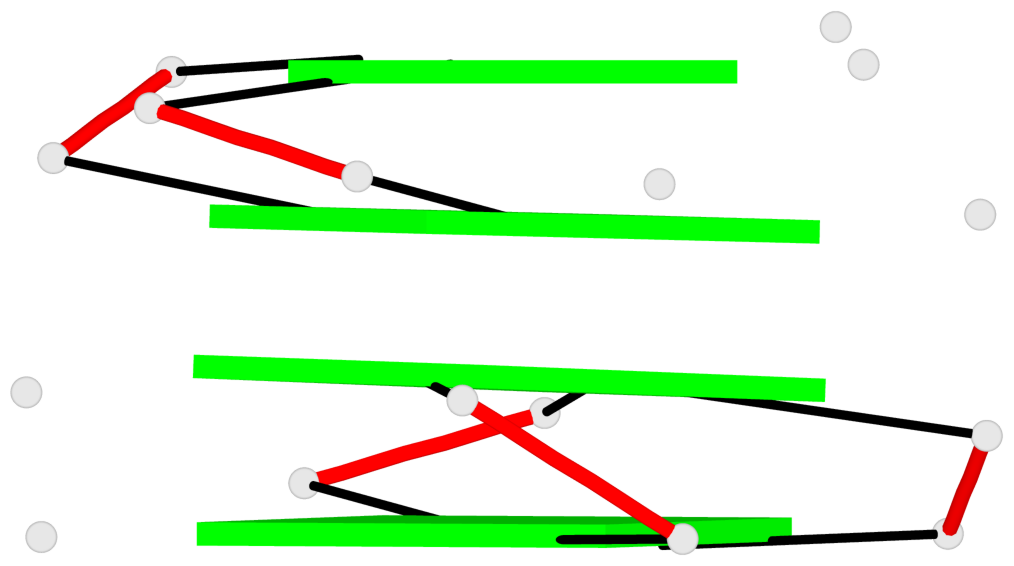
List of 4 non-stem G4-loops (including the two closing Gs)
1 type=V-shaped helix=#1 nts=4 GTGG A.DG15,A.DT16,A.DG17,A.DG18 2* type=V-shaped helix=#1 nts=4 GGTG A.DG23,A.DG24,A.DT25,A.DG26 3 type=V-shaped helix=#2 nts=4 GTGG B.DG15,B.DT16,B.DG17,B.DG18 4* type=V-shaped helix=#2 nts=4 GGTG B.DG23,B.DG24,B.DT25,B.DG26