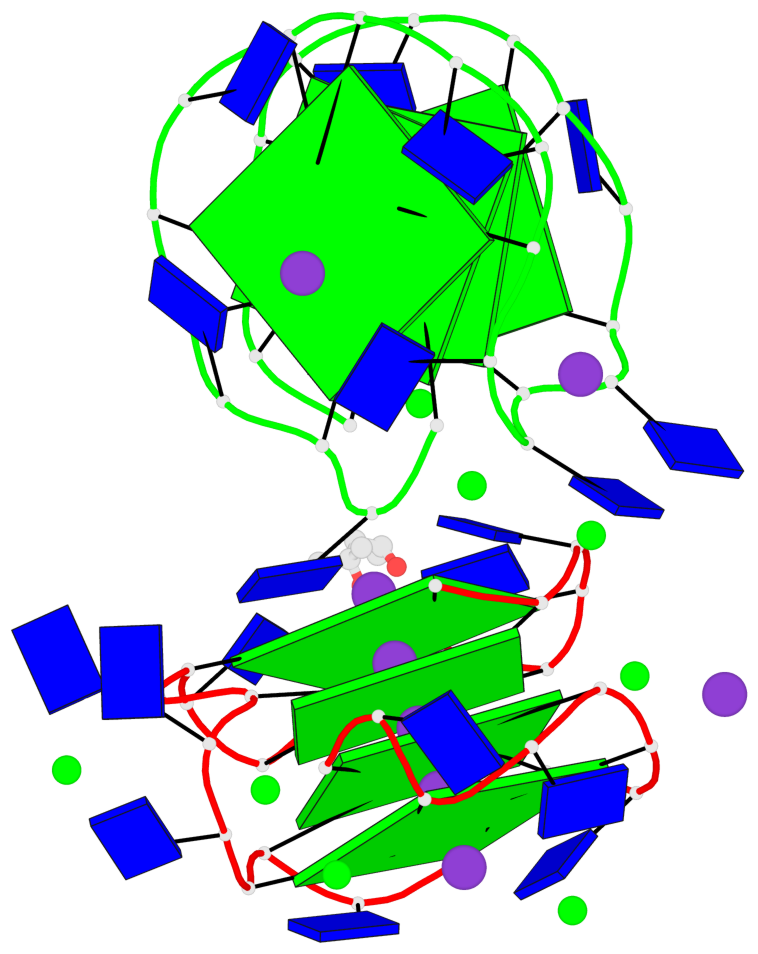
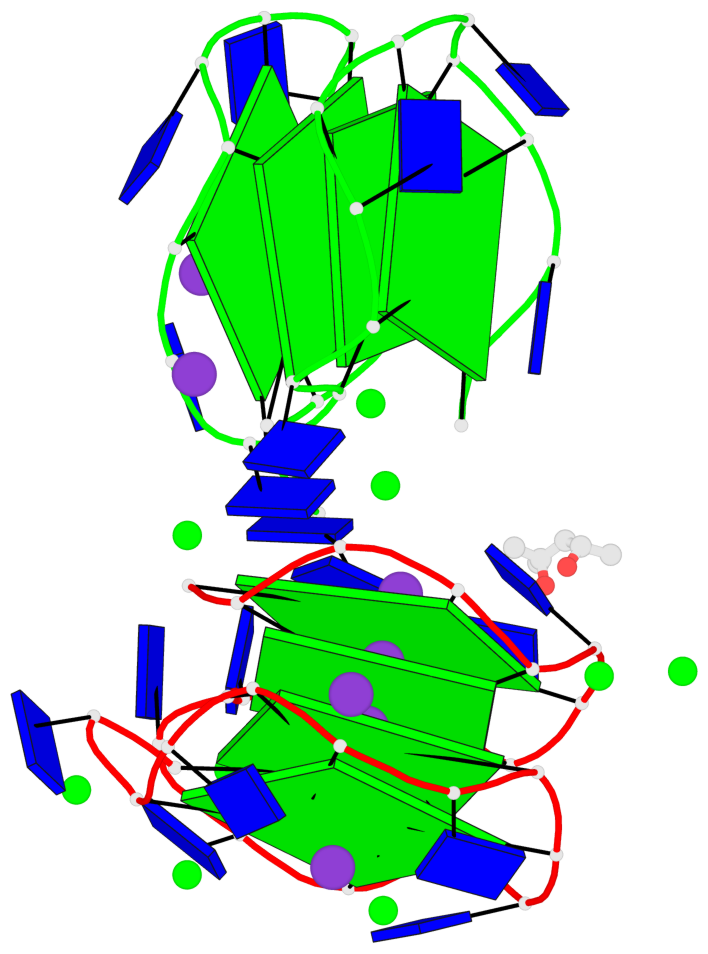
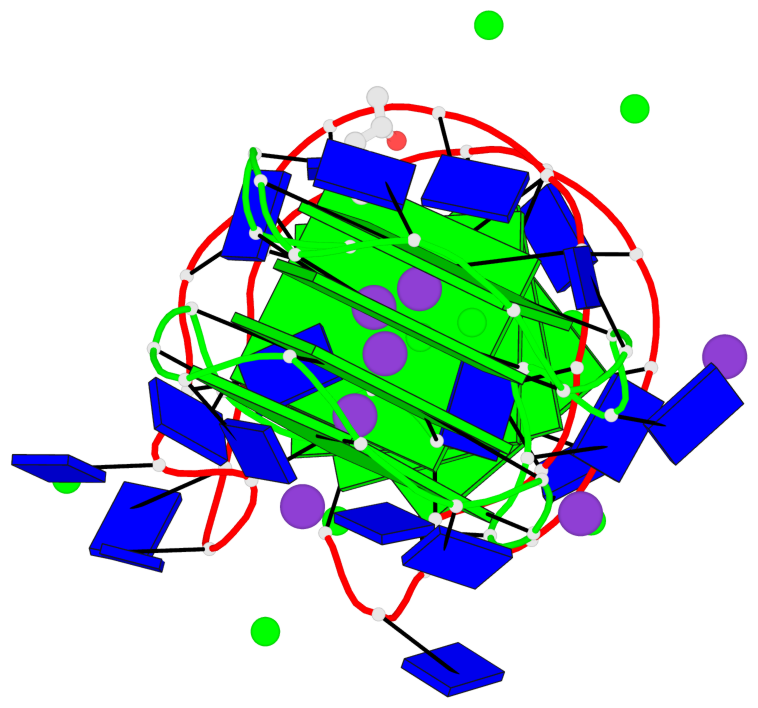
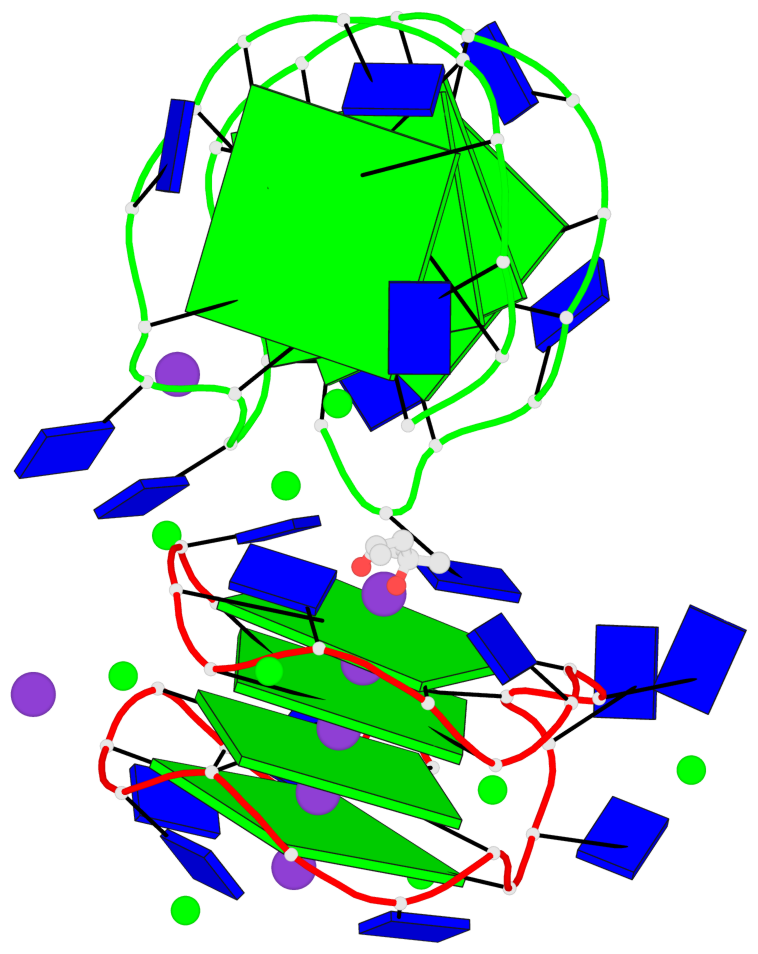
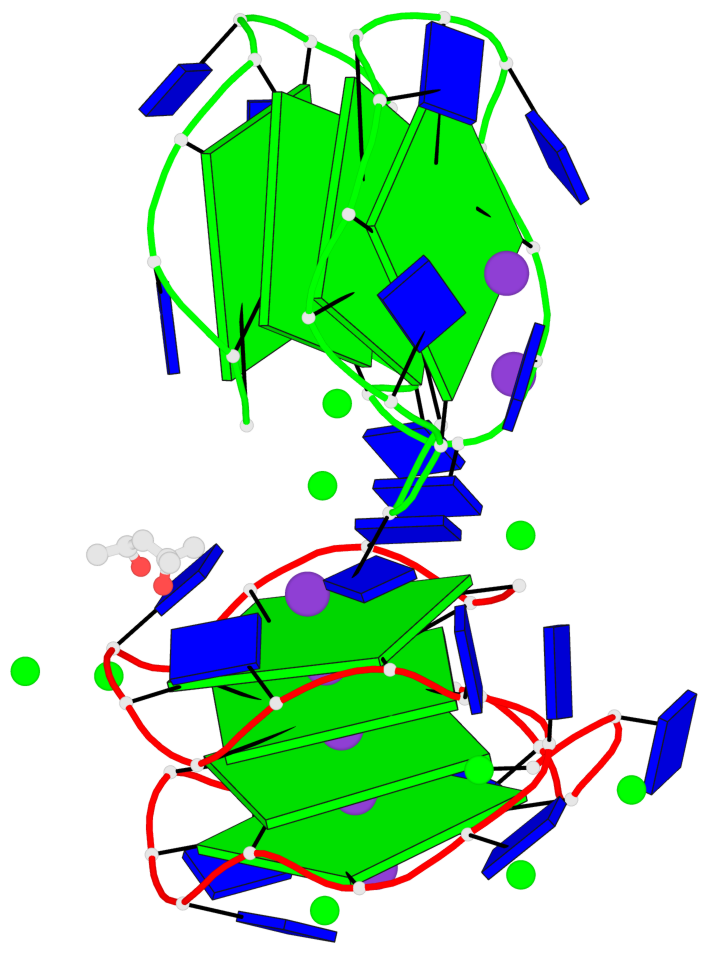
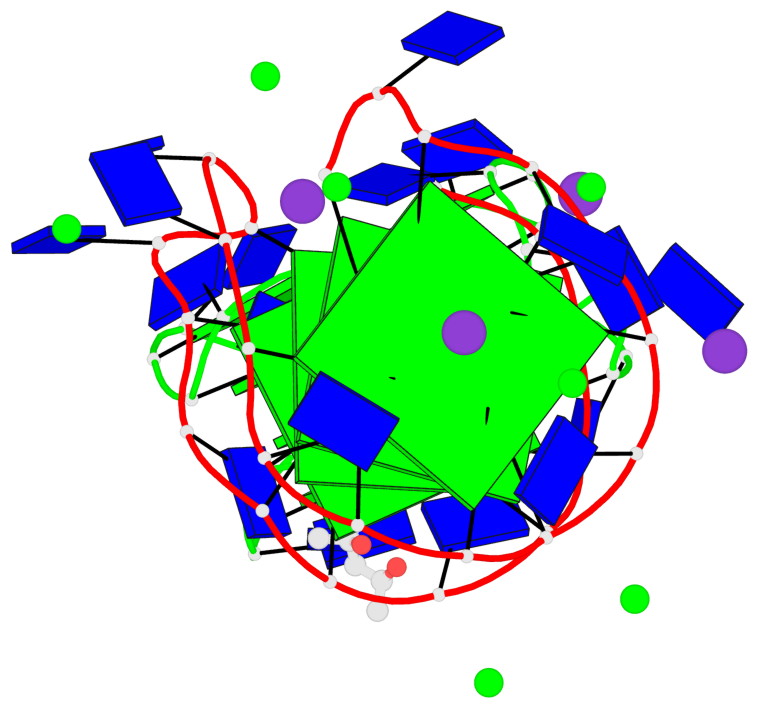
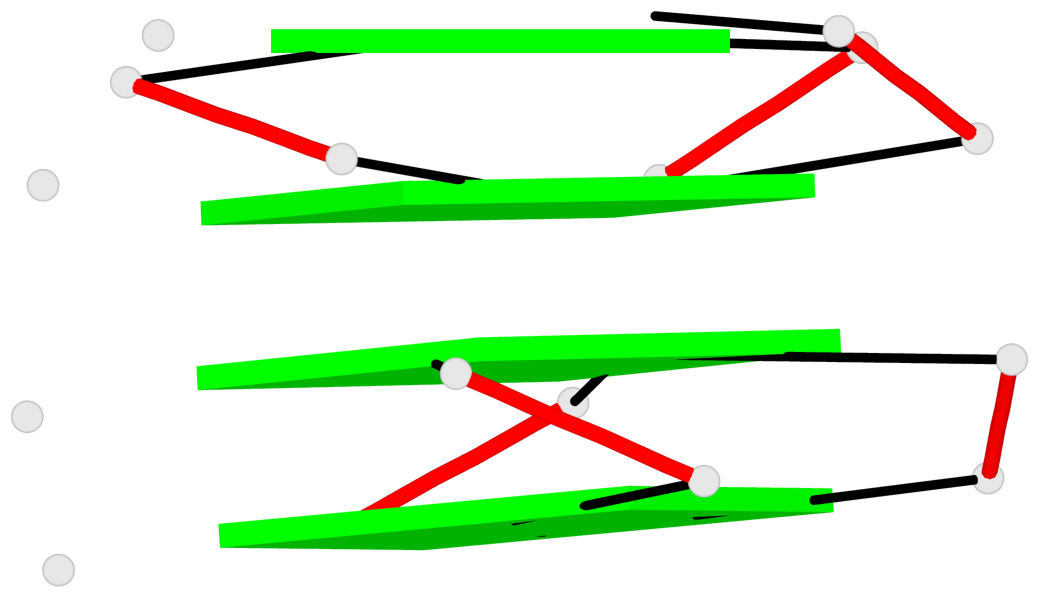
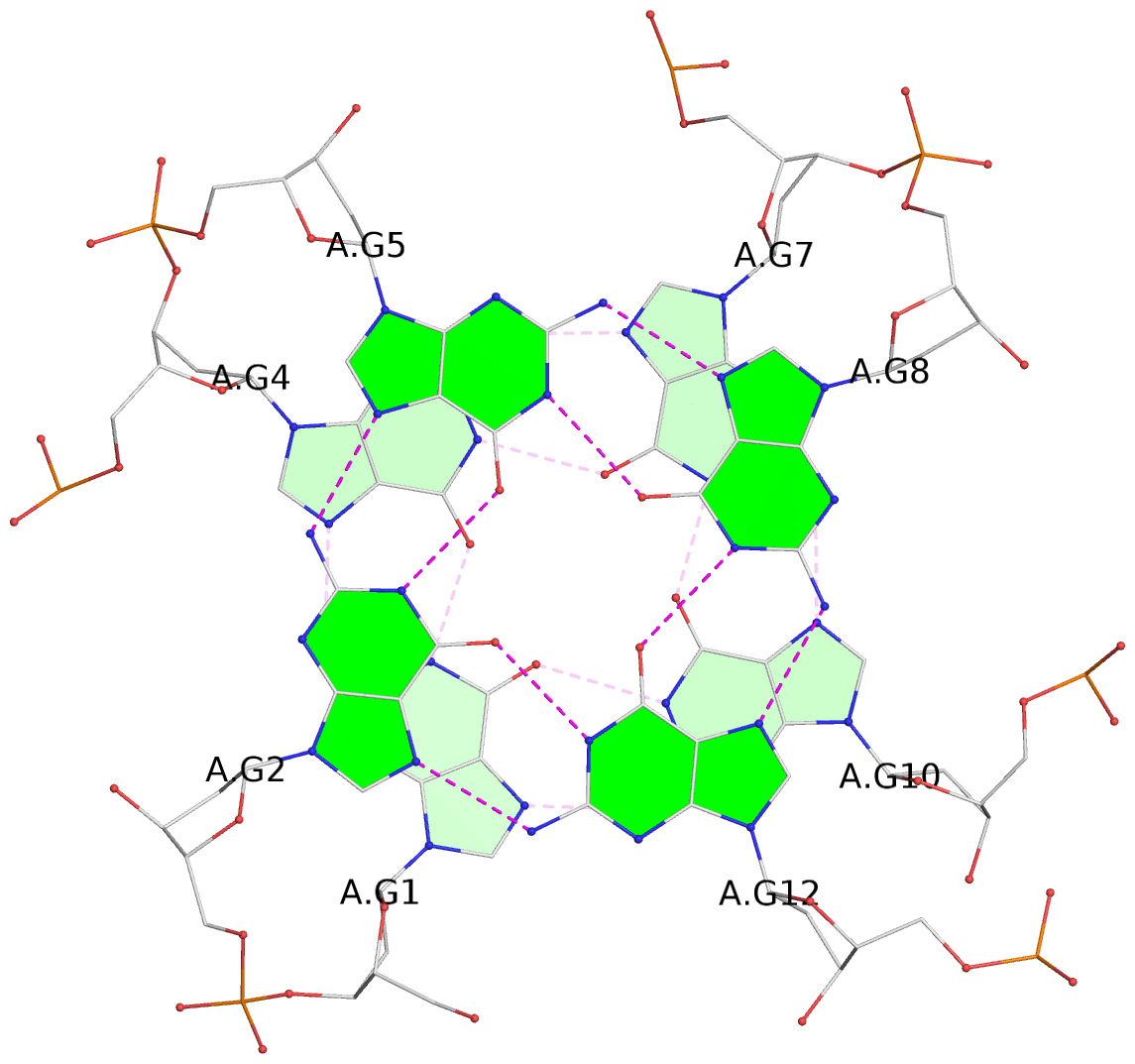
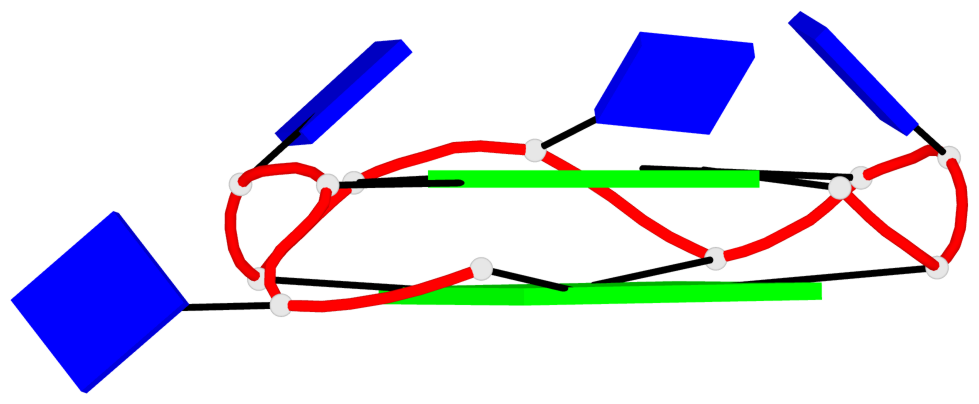
Detailed DSSR results for the G-quadruplex: PDB entry 7dfy
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7dfy
- Class
- DNA
- Method
- X-ray (1.69 Å)
- Summary
- Novel motif for left-handed g-quadruplex formation
- Reference
- Das P, Winnerdy FR, Maity A, Mechulam Y, Phan AT (2021): "A novel minimal motif for left-handed G-quadruplex formation." Chem.Commun.(Camb.), 57, 2527-2530. doi: 10.1039/d0cc08146a.
- Abstract
- A recent study on the left-handed G-quadruplex (LHG4) DNA revealed a 12-nt minimal motif GTGGTGGTGGTG with the ability to independently form an LHG4 and to drive an adjacent sequence to LHG4 formation. Here we have identified a second LHG4-forming motif, GGTGGTGGTGTG, and determined the X-ray crystal structure of an LHG4 involving this motif. Our structural analysis indicated the role of split guanines and single thymine loops in promoting LHG4 formation.
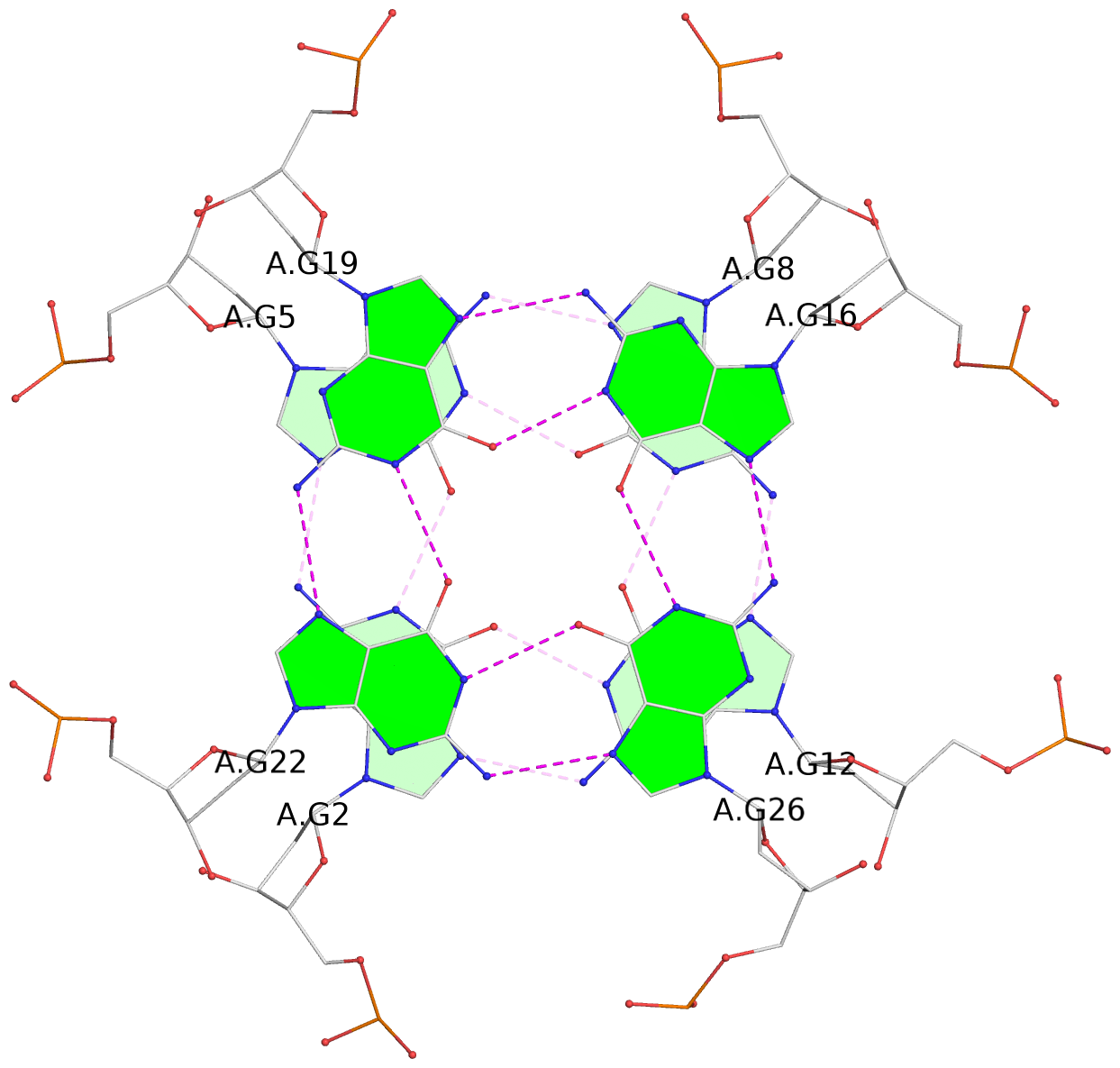
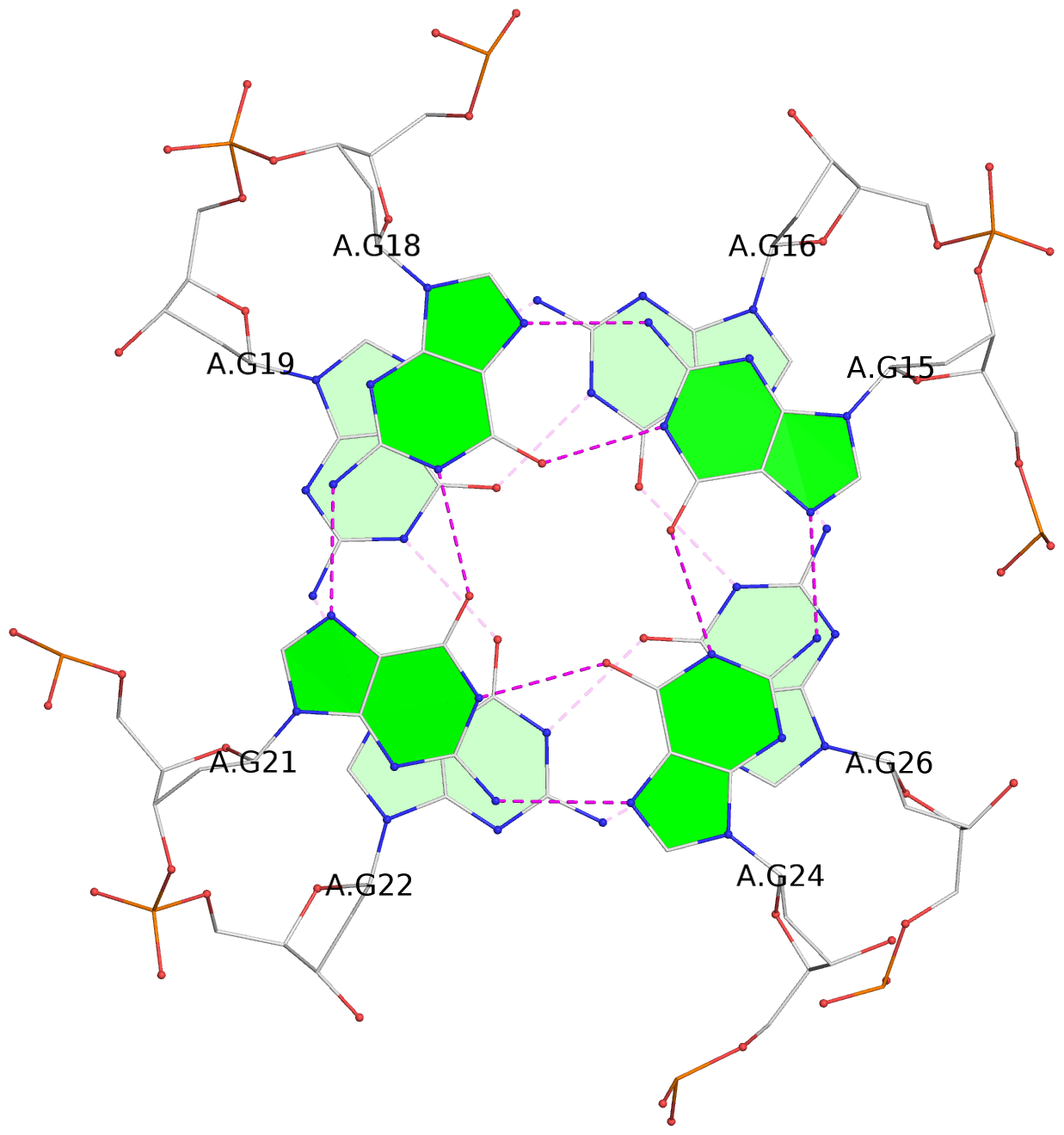
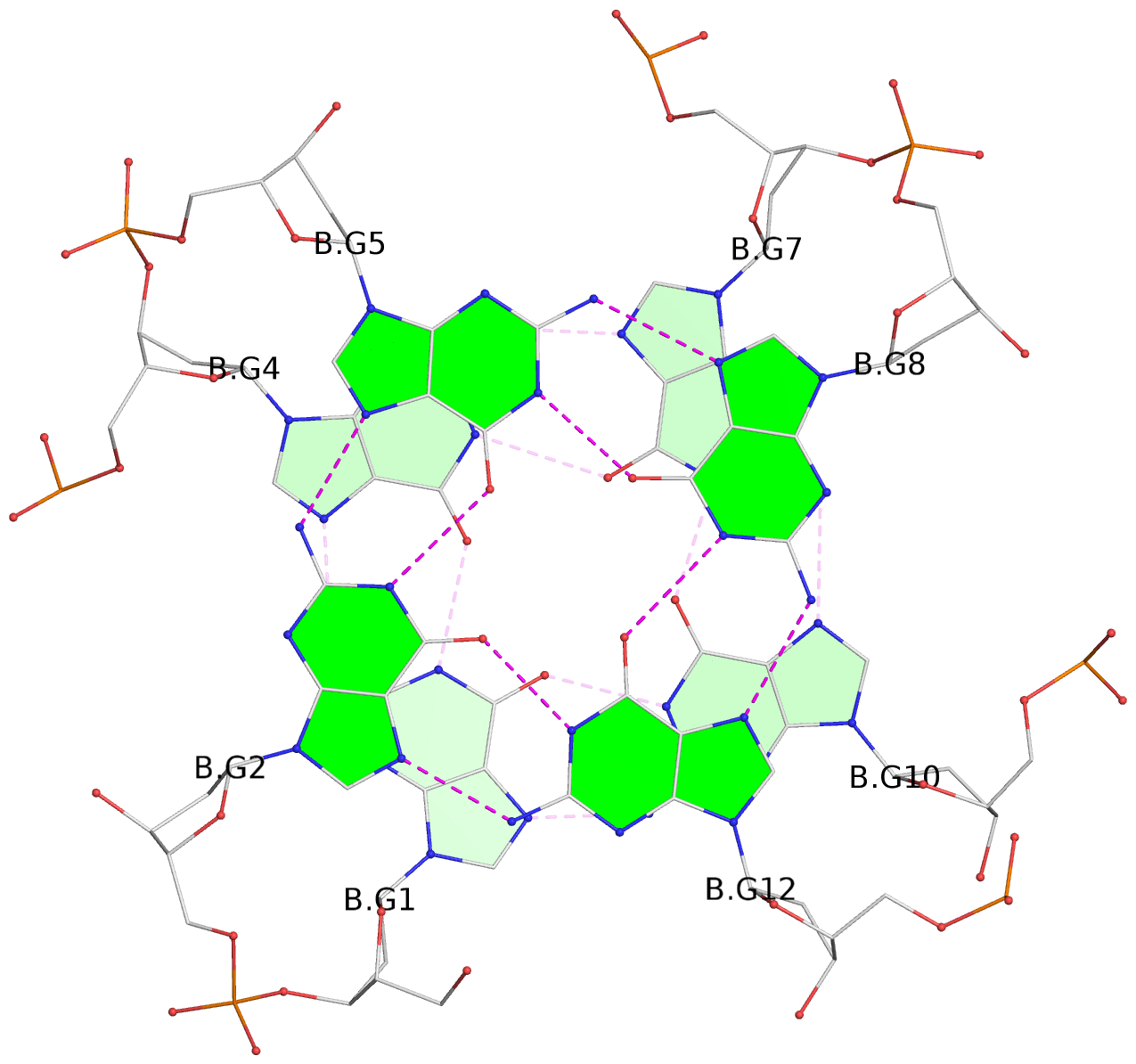
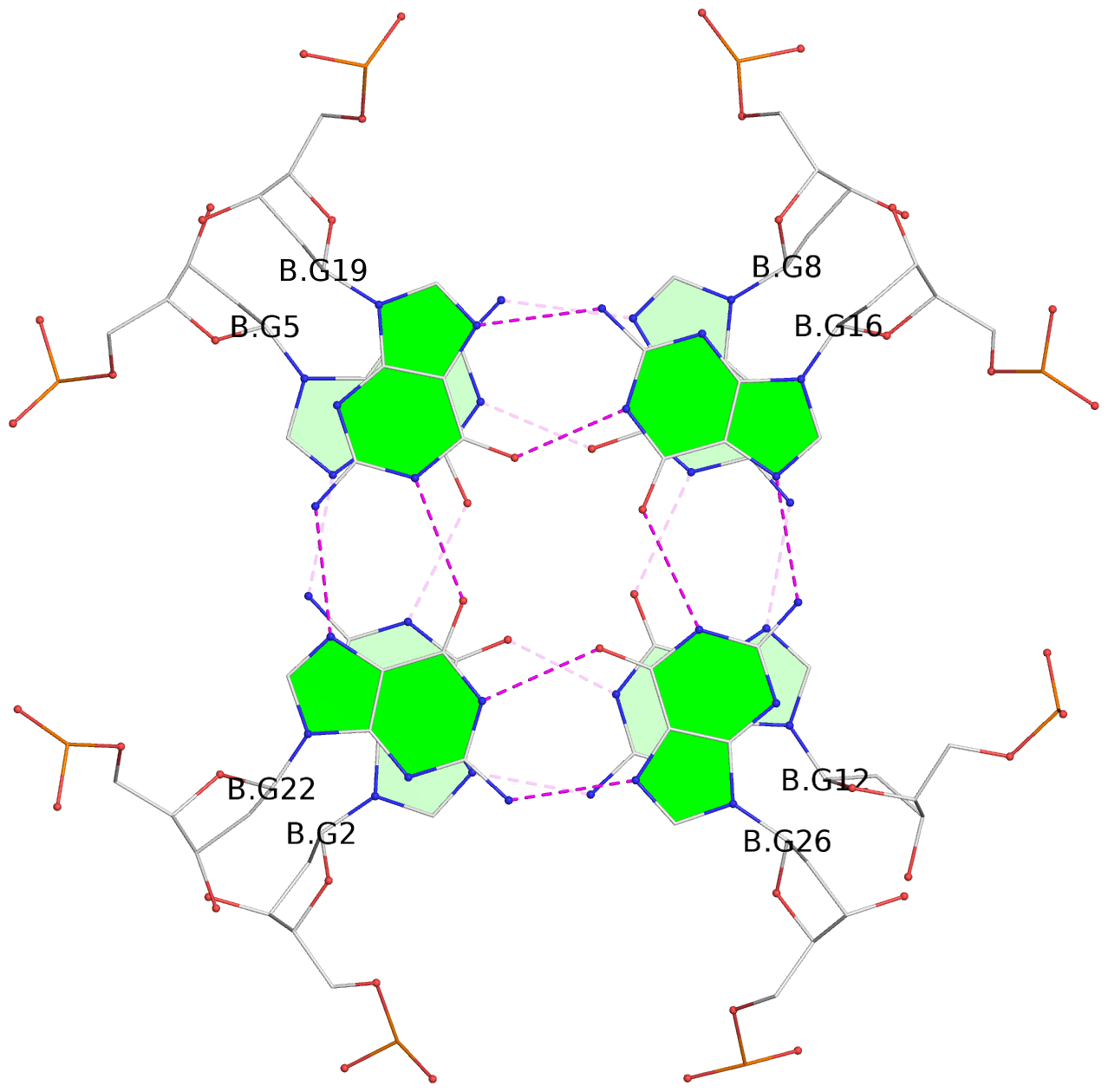
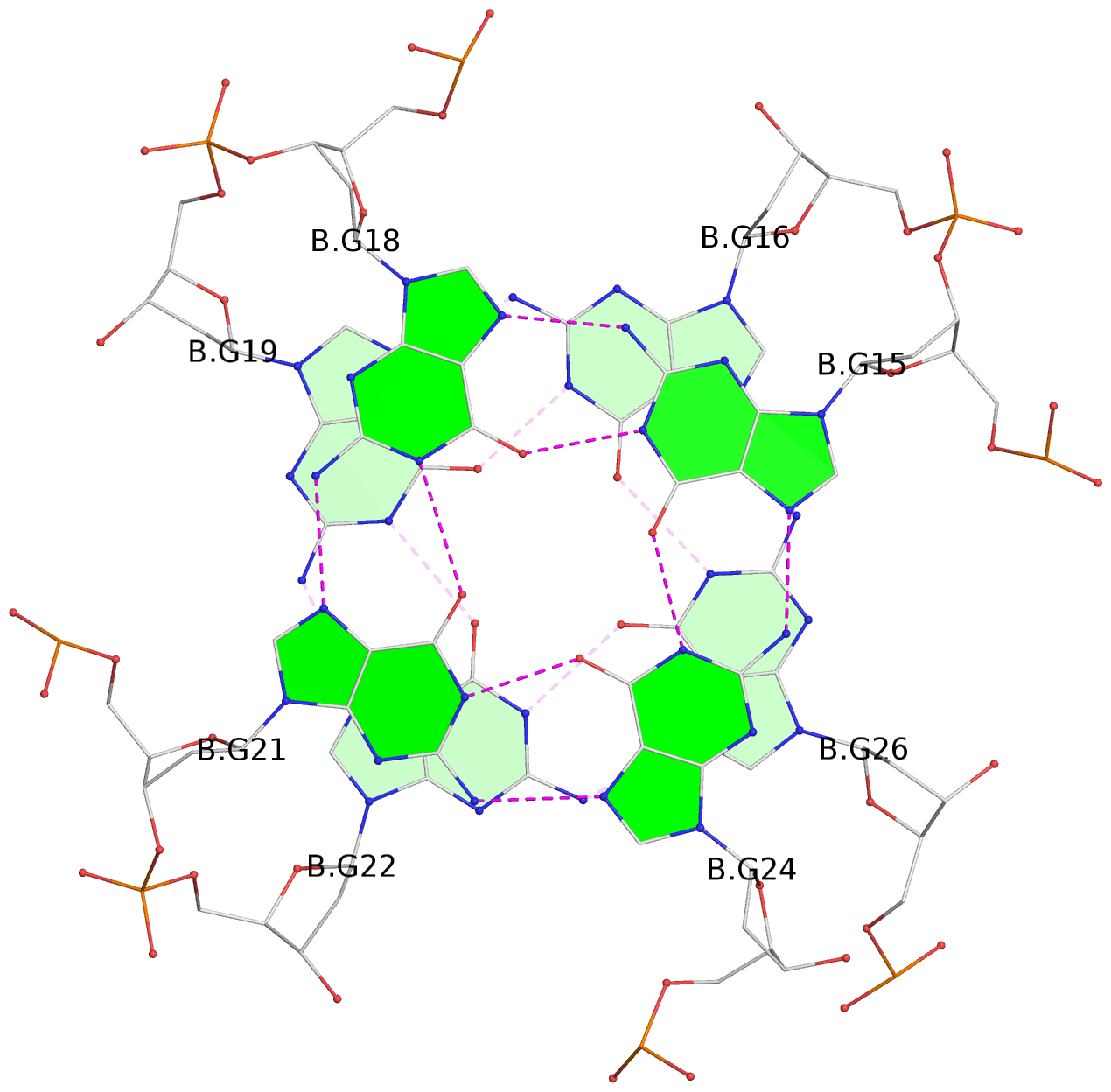
- G4 notes
- 8 G-tetrads, 2 G4 helices, 4 G4 stems, 2 G4 coaxial stacks, 2(+P+P+P), parallel(4+0), UUUU, coaxial interfaces: 3'/3', negative twist
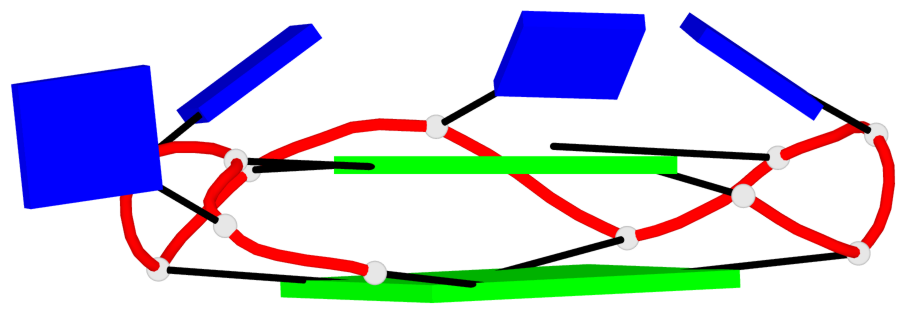
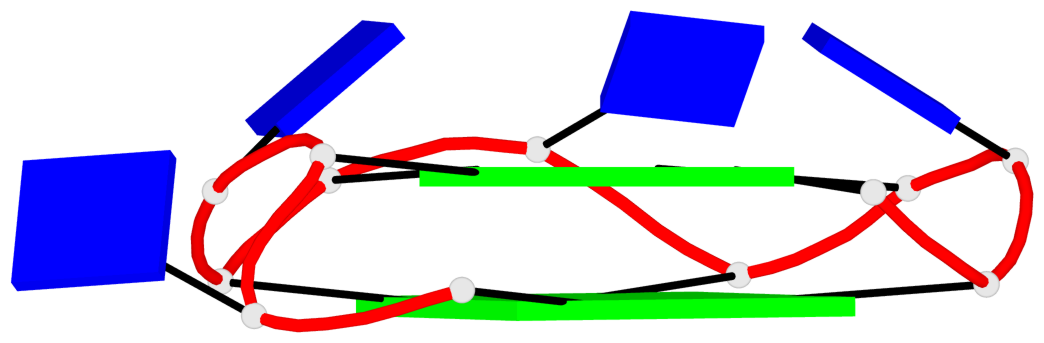
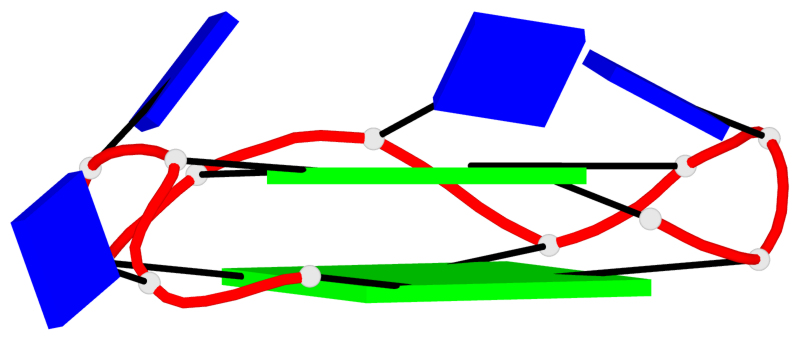
Base-block schematics in six views
List of 8 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.224 type=other nts=4 GGGG A.DG1,A.DG4,A.DG7,A.DG10 2 glyco-bond=---- sugar=---- groove=---- planarity=0.096 type=planar nts=4 GGGG A.DG2,A.DG5,A.DG8,A.DG12 3 glyco-bond=---- sugar=---- groove=---- planarity=0.204 type=other nts=4 GGGG A.DG15,A.DG18,A.DG21,A.DG24 4 glyco-bond=---- sugar=---- groove=---- planarity=0.086 type=planar nts=4 GGGG A.DG16,A.DG19,A.DG22,A.DG26 5 glyco-bond=---- sugar=---- groove=---- planarity=0.246 type=other nts=4 GGGG B.DG1,B.DG4,B.DG7,B.DG10 6 glyco-bond=---- sugar=---- groove=---- planarity=0.147 type=planar nts=4 GGGG B.DG2,B.DG5,B.DG8,B.DG12 7 glyco-bond=---- sugar=---- groove=---- planarity=0.181 type=other nts=4 GGGG B.DG15,B.DG18,B.DG21,B.DG24 8 glyco-bond=---- sugar=---- groove=---- planarity=0.100 type=planar nts=4 GGGG B.DG16,B.DG19,B.DG22,B.DG26
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 2 stems
Helix#2, 4 G-tetrad layers, INTRA-molecular, with 2 stems
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(+P+P+P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(+P+P+P), parallel(4+0)
Stem#3, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(+P+P+P), parallel(4+0)
Stem#4, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(+P+P+P), parallel(4+0)
List of 2 G4 coaxial stacks
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [3'/3'] 2 G4 helix#2 contains 2 G4 stems: [#3,#4] [3'/3']