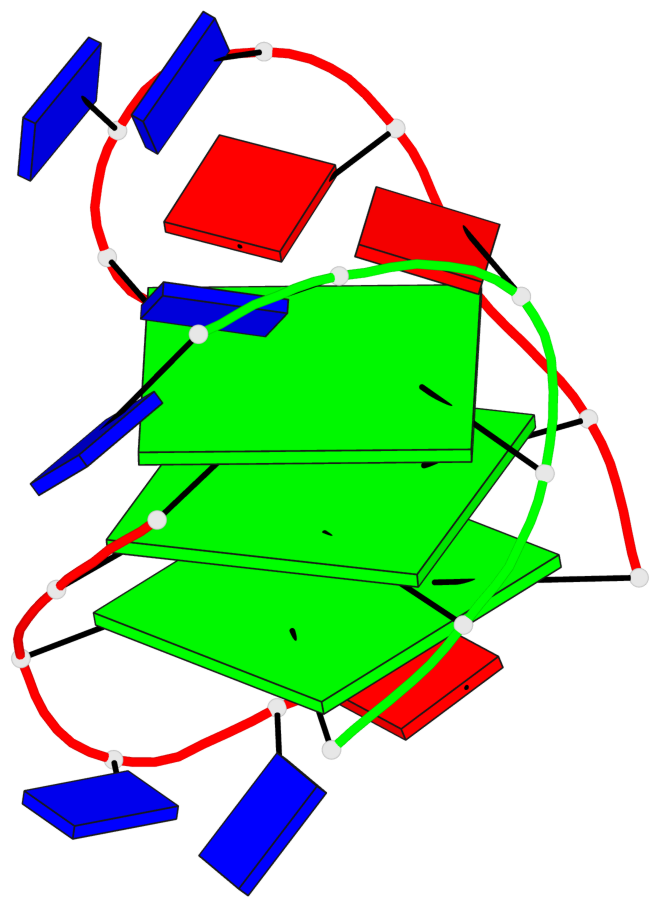
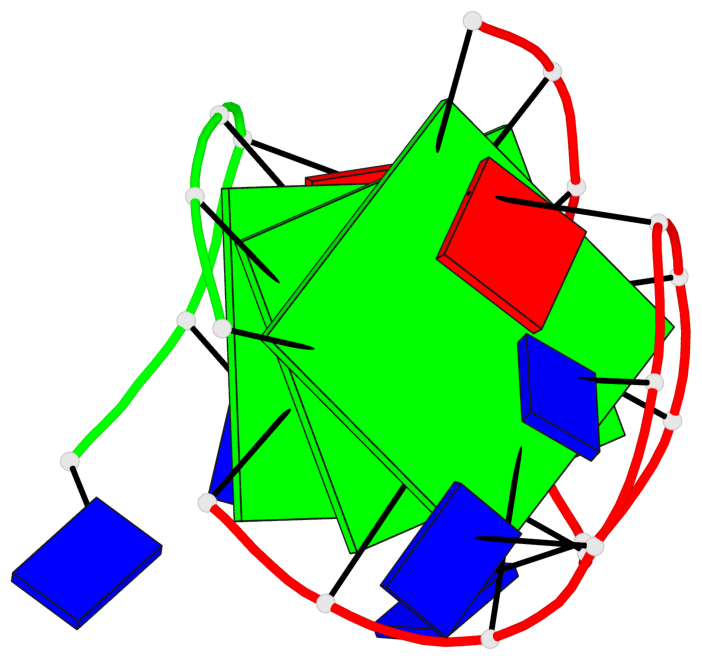
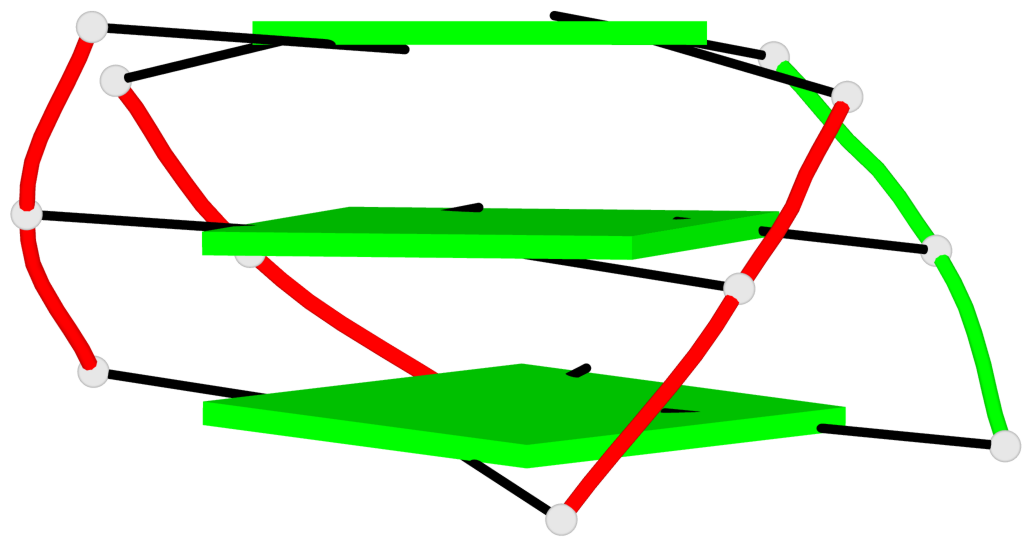
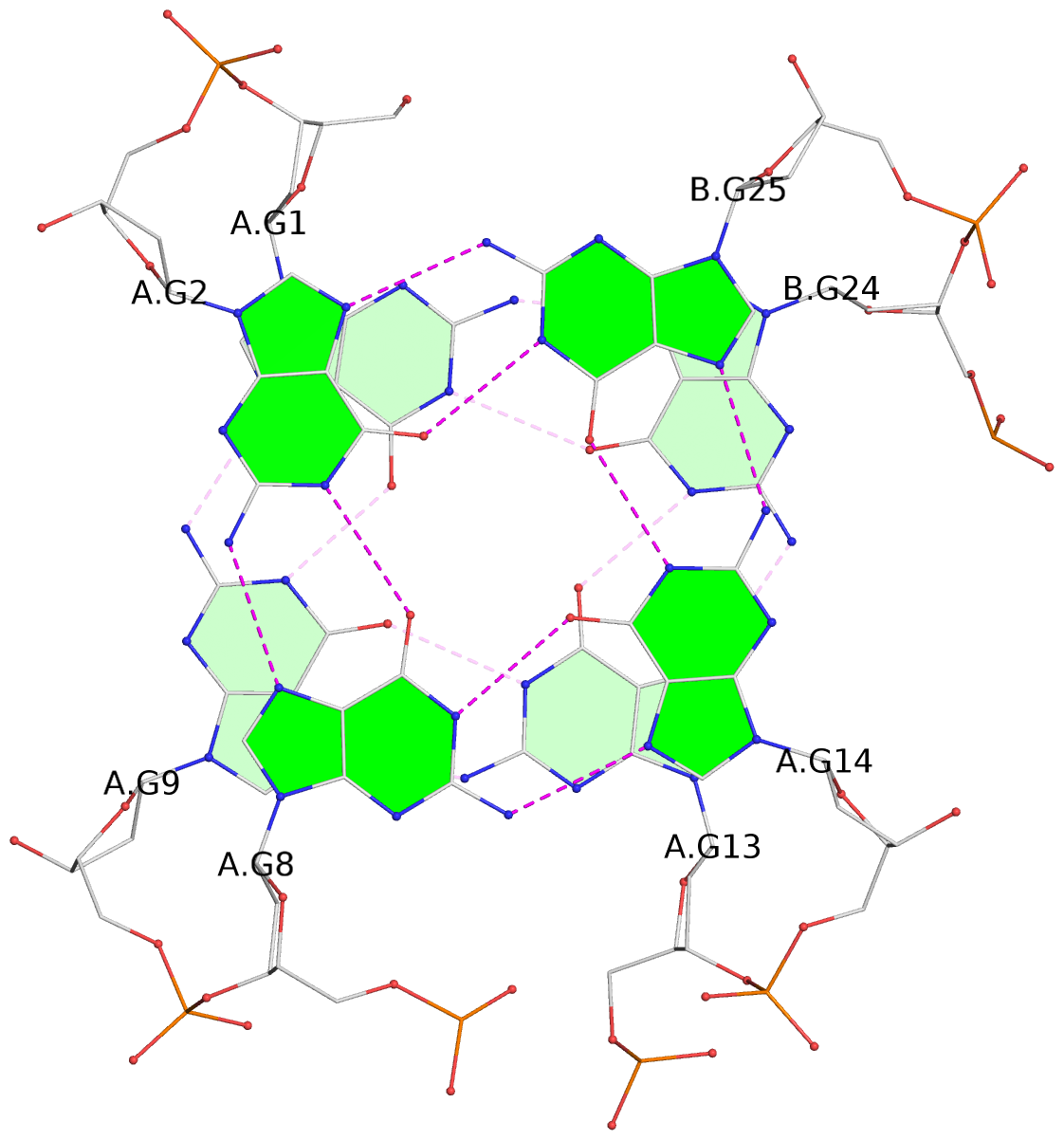
Detailed DSSR results for the G-quadruplex: PDB entry 7do1
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7do1
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of a heteromolecular telomeric (3+1) g-quadruplex containing right loop progression
- Reference
- Fu W, Jing H, Xu X, Xu S, Wang T, Hu W, Li H, Zhang N (2021): "Two coexisting pseudo-mirror heteromolecular telomeric G-quadruplexes in opposite loop progressions differentially recognized by a low equivalent of Thioflavin T." Nucleic Acids Res., 49, 10717-10734. doi: 10.1093/nar/gkab755.
- Abstract
- The final 3'-terminal residue of the telomeric DNA G-overhang is inherently less precise. Here, we describe how alteration of the last 3'-terminal base affects the mutual recognition between two different G-rich oligomers of human telomeric DNA in the formation of heteromolecular G-quadruplexes (hetero-GQs). Associations between three- and single-repeat fragments of human telomeric DNA, target d(GGGTTAGGGTTAGGG) and probe d(TAGGGT), in Na+ solution yield two coexisting forms of (3 + 1) hybrid hetero-GQs: the kinetically favourable LLP-form (left loop progression) and the thermodynamically controlled RLP-form (right loop progression). However, only the adoption of a single LLP-form has been previously reported between the same probe d(TAGGGT) and a target variant d(GGGTTAGGGTTAGGGT) having one extra 3'-end thymine. Moreover, the flanking base alterations of short G-rich probe variants also significantly affect the loop progressions of hetero-GQs. Although seemingly two pseudo-mirror counter partners, the RLP-form exhibits a preference over the LLP-form to be recognized by a low equivalent of fluorescence dye thioflavin T (ThT). To a greater extent, ThT preferentially binds to RLP hetero-GQ than with the corresponding telomeric DNA duplex context or several other representative unimolecular GQs.
- G4 notes
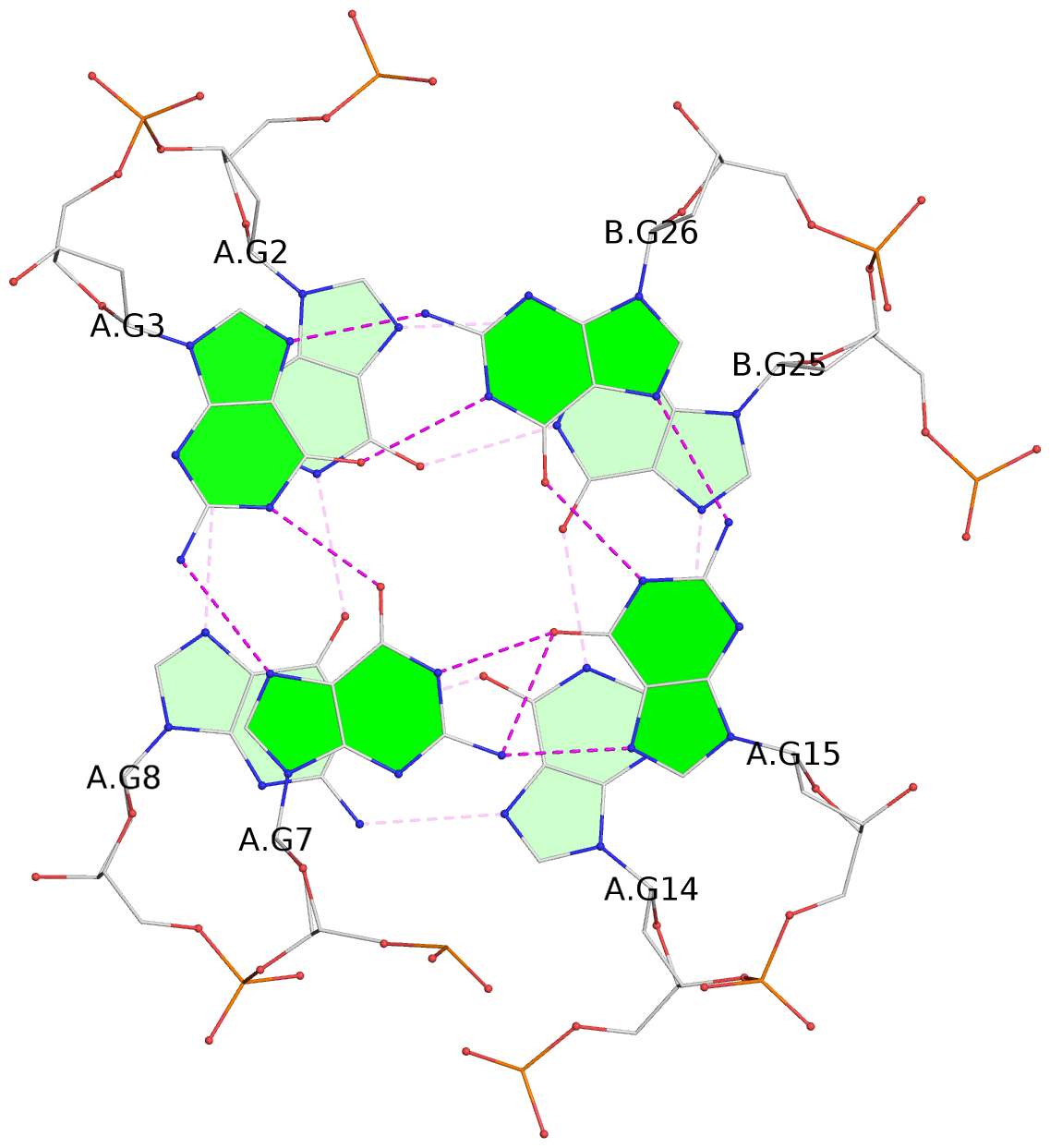
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, (3+1), UDUU
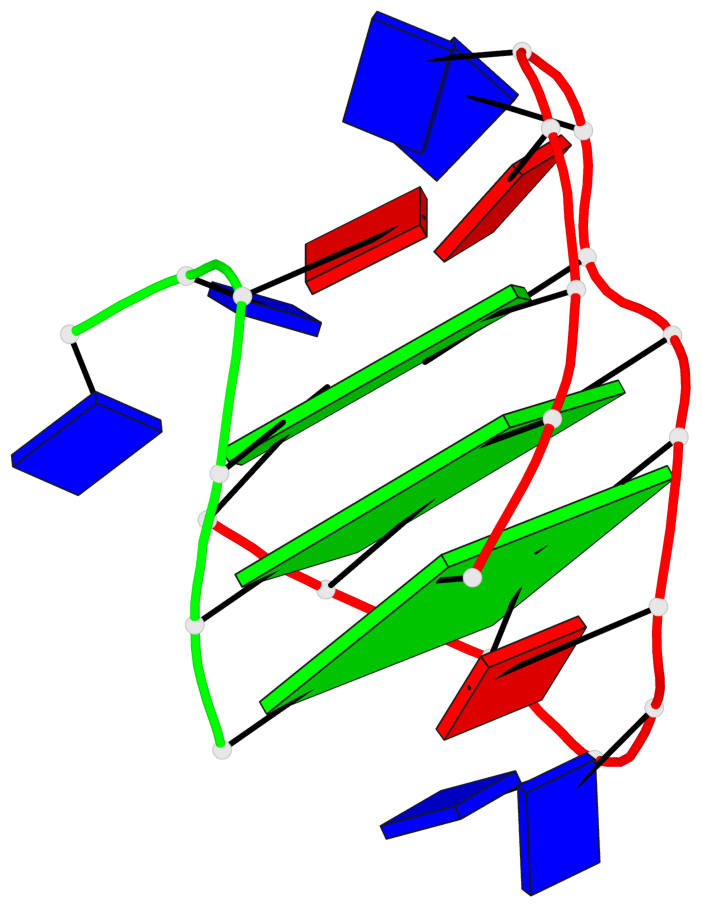
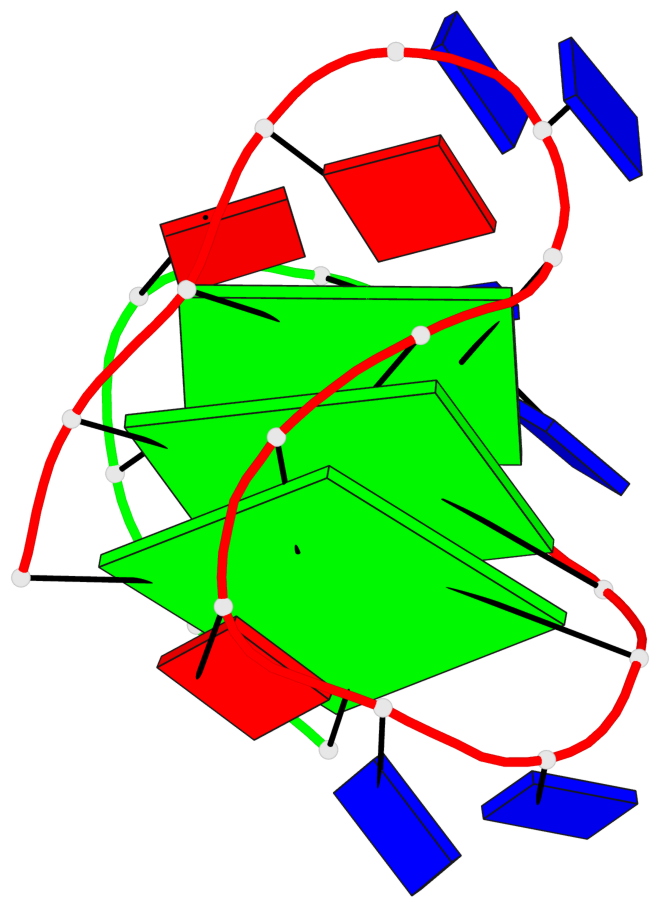
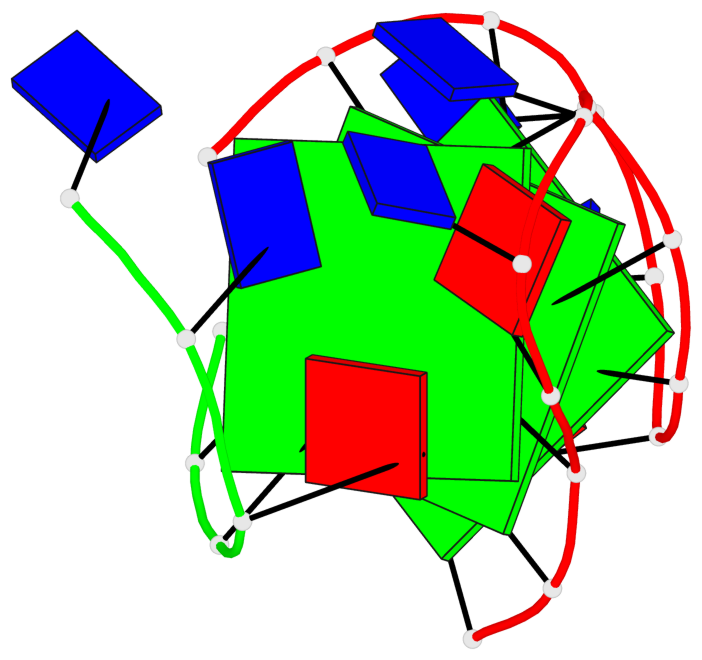
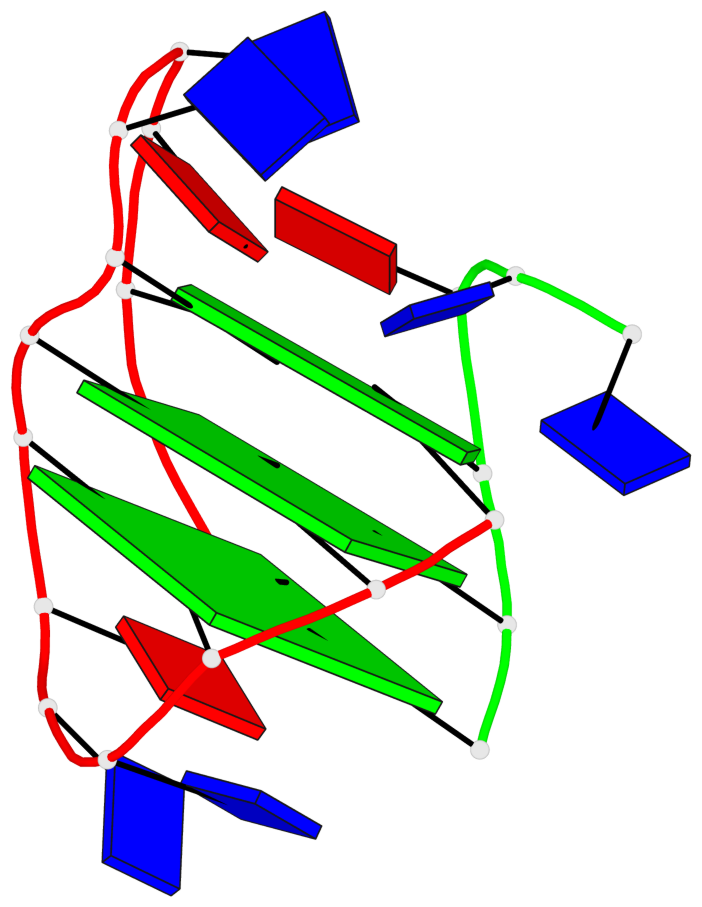
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=---- groove=wn-- planarity=0.159 type=planar nts=4 GGGG A.DG1,A.DG9,A.DG13,B.DG24 2 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.141 type=planar nts=4 GGGG A.DG2,A.DG8,A.DG14,B.DG25 3 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.232 type=other nts=4 GGGG A.DG3,A.DG7,A.DG15,B.DG26
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.