Detailed DSSR results for the G-quadruplex: PDB entry 7e5p
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7e5p
- Class
- DNA
- Method
- NMR
- Summary
- Aptamer enhancing peroxidase activity of myoglobin
- Reference
- Tsukakoshi K, Yamagishi Y, Kanazashi M, Nakama K, Oshikawa D, Savory N, Matsugami A, Hayashi F, Lee J, Saito T, Sode K, Khunathai K, Kuno H, Ikebukuro K (2021): "G-quadruplex-forming aptamer enhances the peroxidase activity of myoglobin against luminol." Nucleic Acids Res., 49, 6069-6081. doi: 10.1093/nar/gkab388.
- Abstract
- Aptamers can control the biological functions of enzymes, thereby facilitating the development of novel biosensors. While aptamers that inhibit catalytic reactions of enzymes were found and used as signal transducers to sense target molecules in biosensors, no aptamers that amplify enzymatic activity have been identified. In this study, we report G-quadruplex (G4)-forming DNA aptamers that upregulate the peroxidase activity in myoglobin specifically for luminol. Using in vitro selection, one G4-forming aptamer that enhanced chemiluminescence from luminol by myoglobin's peroxidase activity was discovered. Through our strategy-in silico maturation, which is a genetic algorithm-aided sequence manipulation method, the enhancing activity of the aptamer was improved by introducing mutations to the aptamer sequences. The best aptamer conserved the parallel G4 property with over 300-times higher luminol chemiluminescence from peroxidase activity more than myoglobin alone at an optimal pH of 5.0. Furthermore, using hemin and hemin-binding aptamers, we demonstrated that the binding property of the G4 aptamers to heme in myoglobin might be necessary to exert the enhancing effect. Structure determination for one of the aptamers revealed a parallel-type G4 structure with propeller-like loops, which might be useful for a rational design of aptasensors utilizing the G4 aptamer-myoglobin pair.
- G4 notes
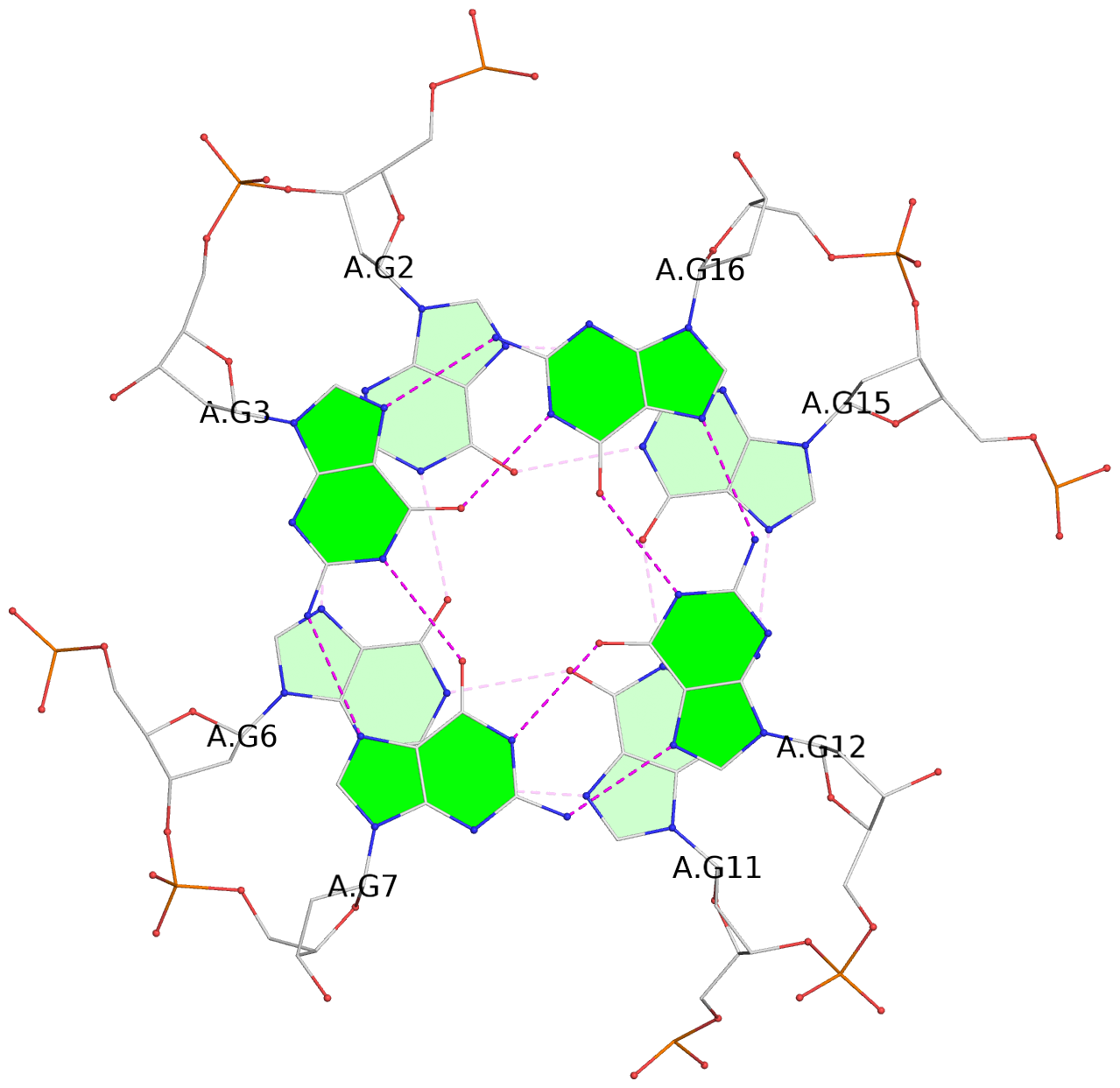
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
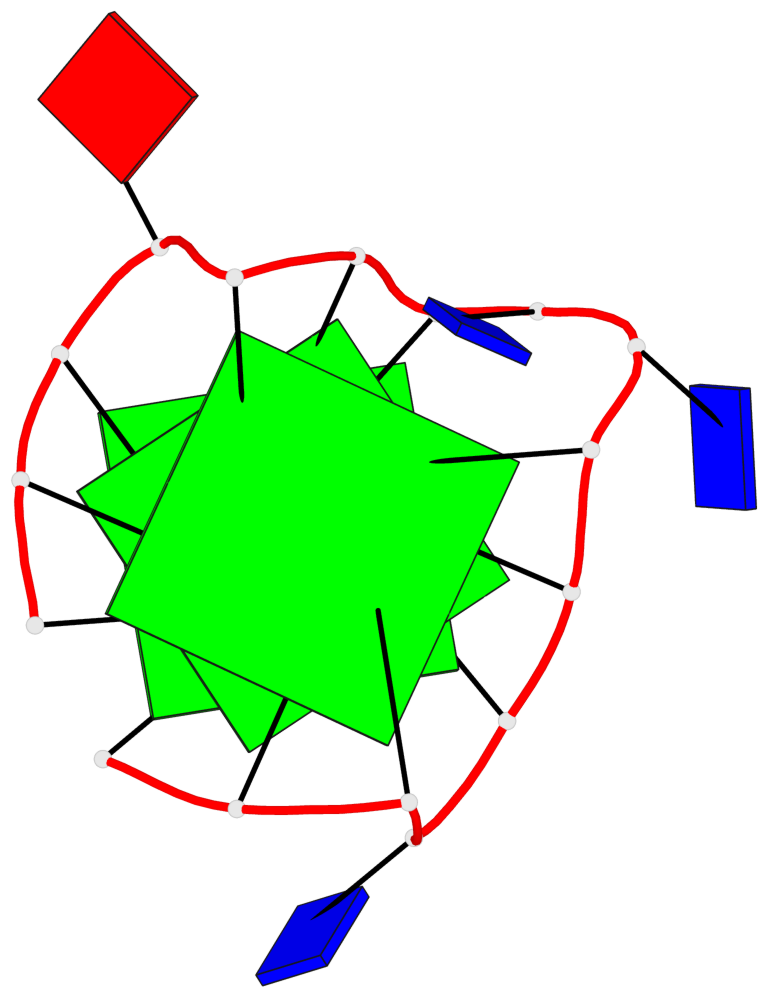
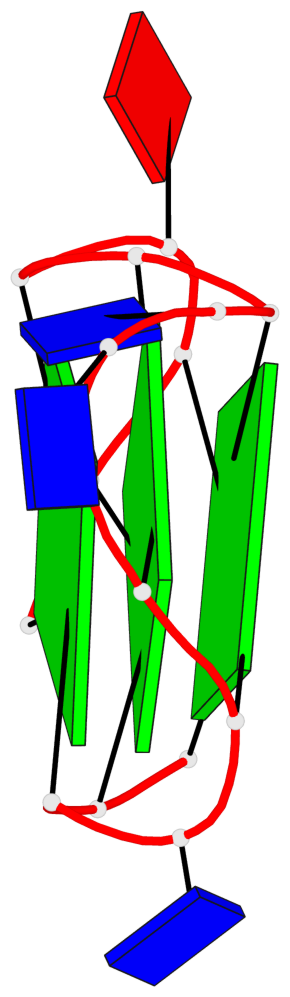
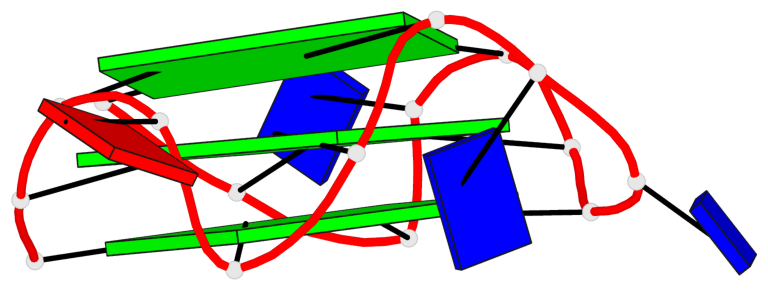
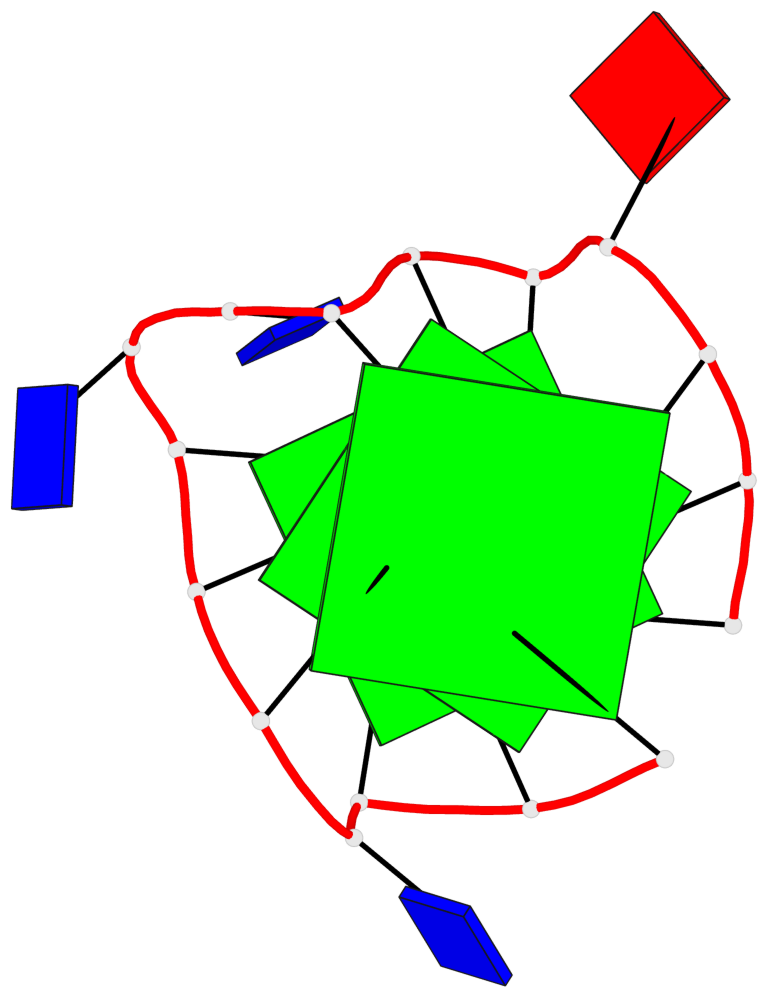
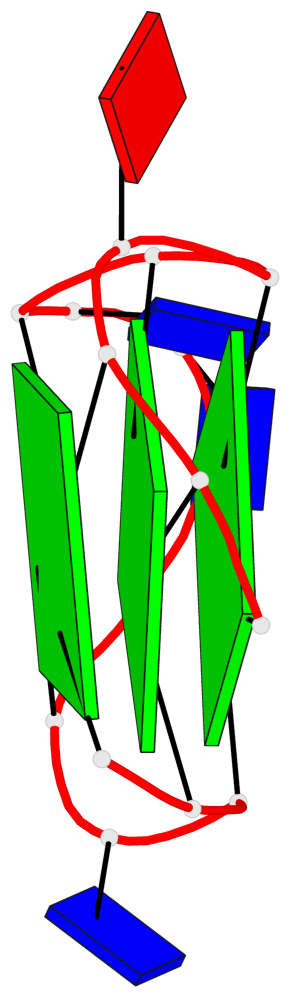
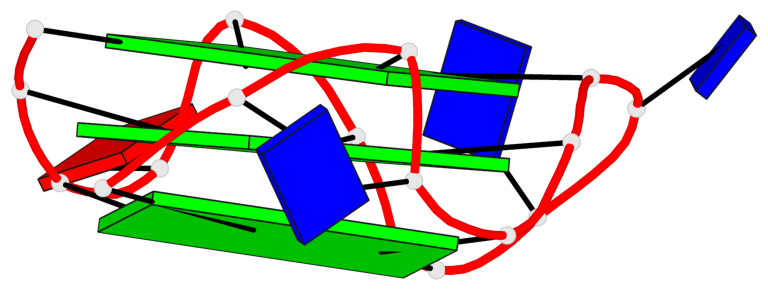
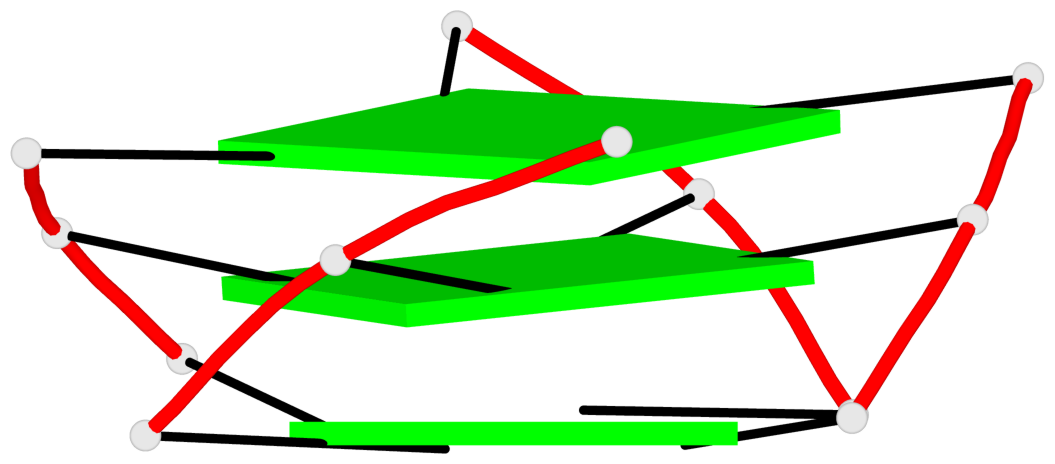
Base-block schematics in six views
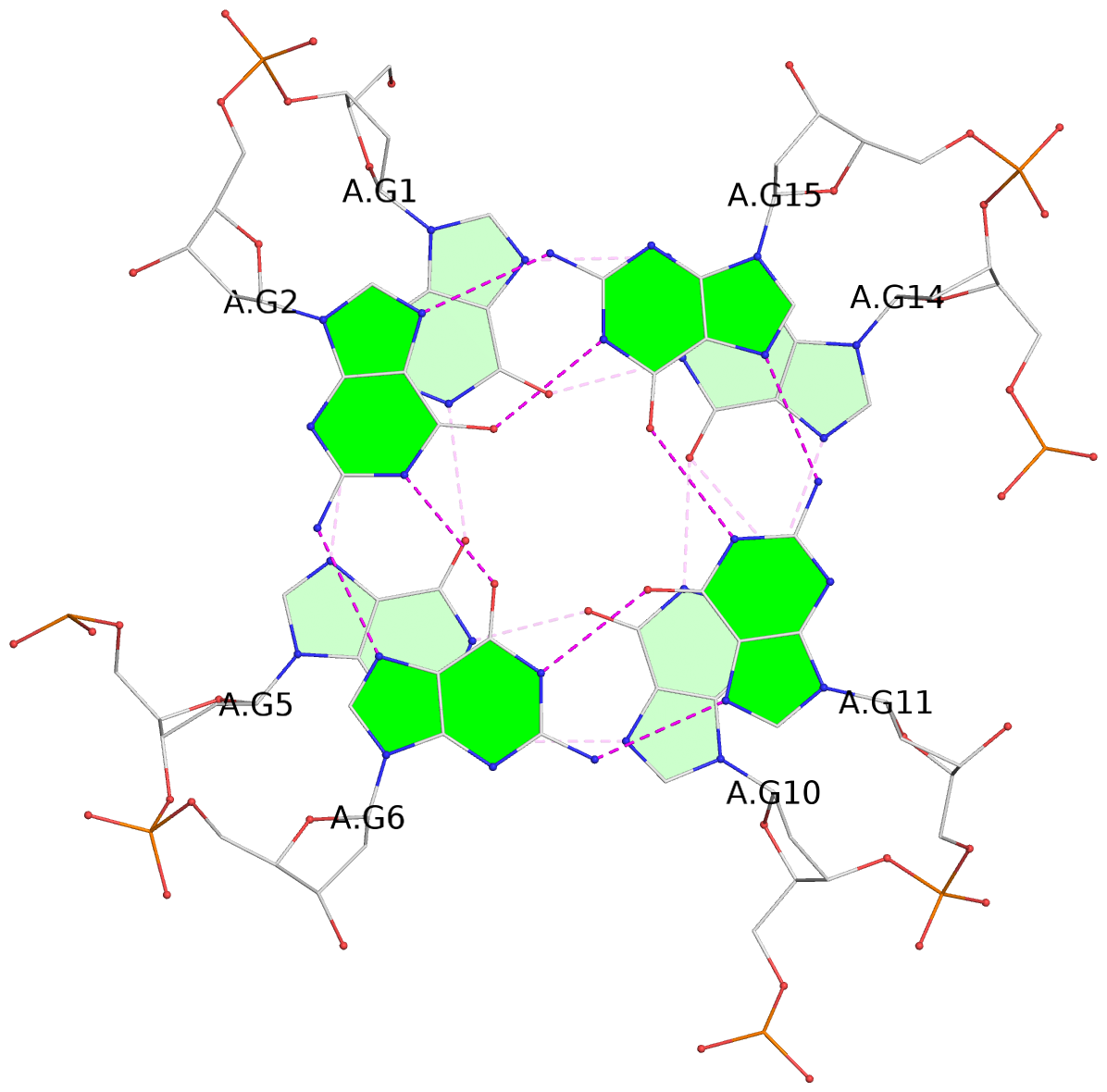
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.216 type=other nts=4 GGGG A.DG1,A.DG5,A.DG10,A.DG14 2 glyco-bond=---- sugar=---- groove=---- planarity=0.124 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG11,A.DG15 3 glyco-bond=---- sugar=---- groove=---- planarity=0.146 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG12,A.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.