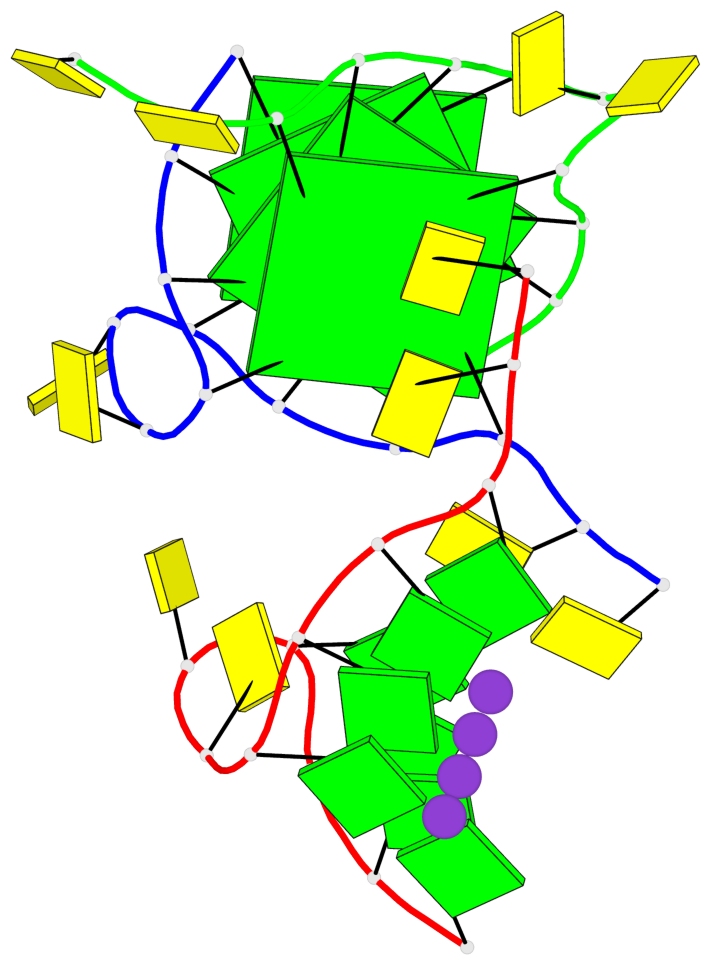
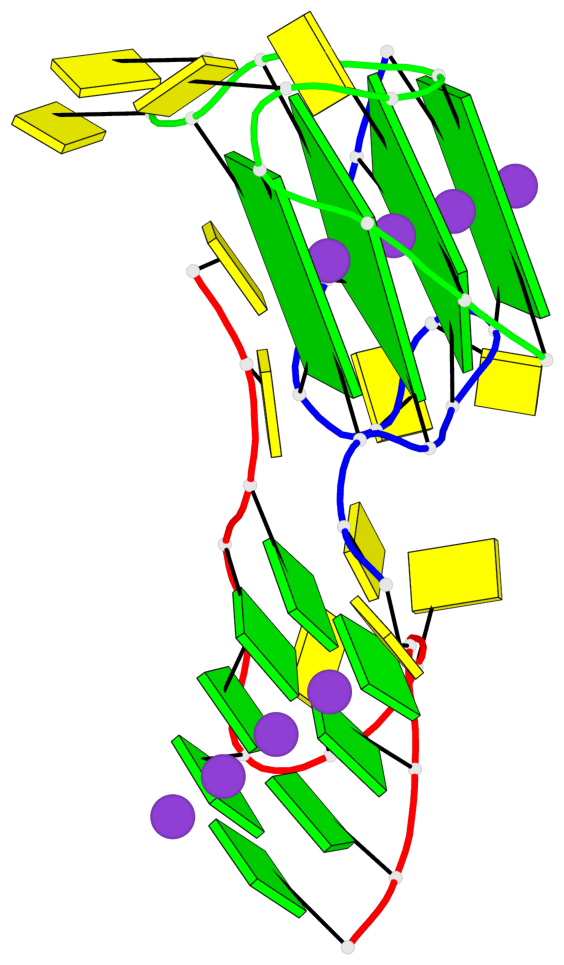
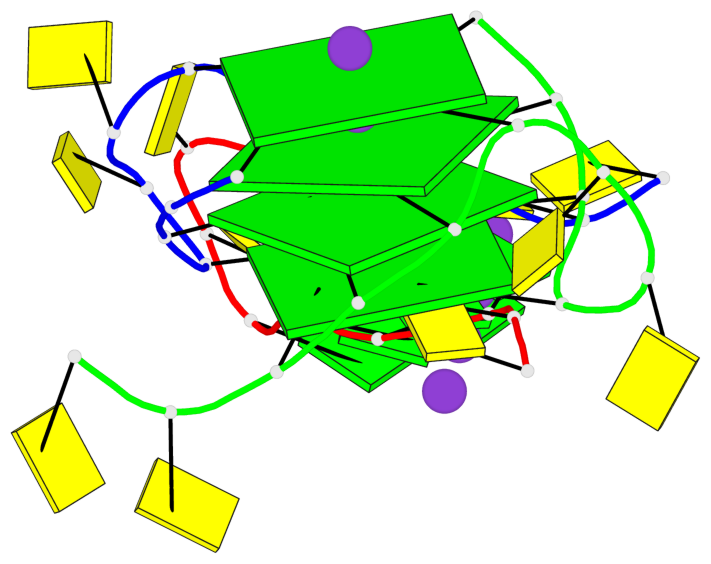
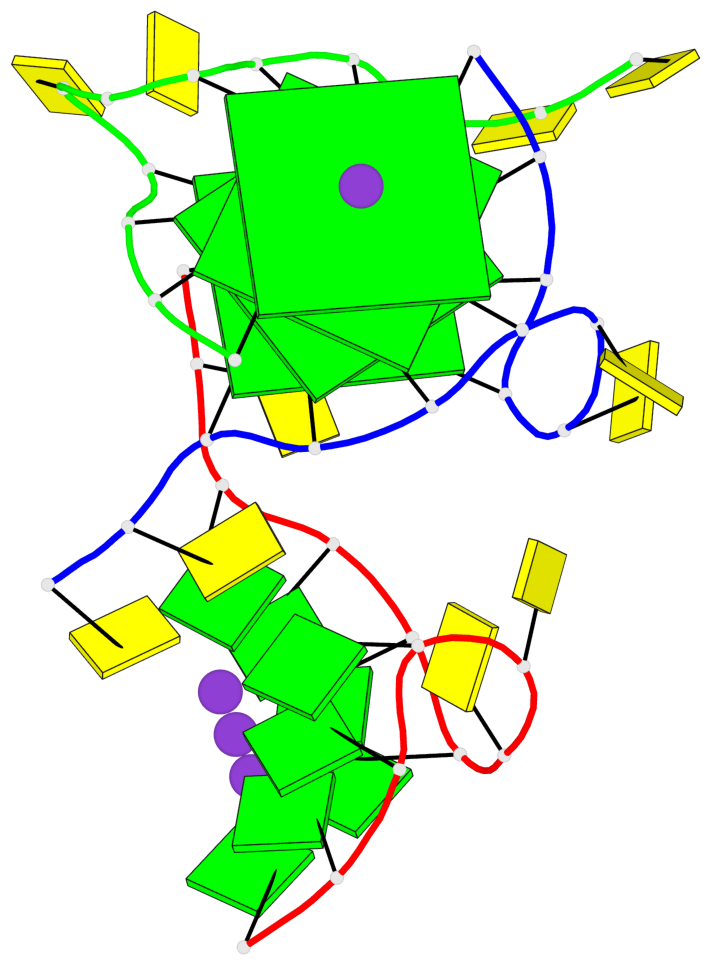
Detailed DSSR results for the G-quadruplex: PDB entry 7ech
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7ech
- Class
- DNA
- Method
- X-ray (2.38 Å)
- Summary
- Crystal structure of d(g4c2)2-k in f222 space group
- Reference
- Geng Y, Liu C, Cai Q, Luo Z, Miao H, Shi X, Xu N, Fung CP, Choy TT, Yan B, Li N, Qian P, Zhou B, Zhu G (2021): "Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence." Nucleic Acids Res., 49, 5881-5890. doi: 10.1093/nar/gkab302.
- Abstract
- The hexanucleotide repeat expansion, GGGGCC (G4C2), within the first intron of the C9orf72 gene is known to be the most common genetic cause of both amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The G4C2 repeat expansions, either DNA or RNA, are able to form G-quadruplexes which induce toxicity leading to ALS/FTD. Herein, we report a novel crystal structure of d(G4C2)2 that self-associates to form an eight-layer parallel tetrameric G-quadruplex. Two d(G4C2)2 associate together as a parallel dimeric G-quadruplex which folds into a tetramer via 5'-to-5' arrangements. Each dimer consists of four G-tetrads connected by two CC propeller loops. Especially, the 3'-end cytosines protrude out and form C·C+•C·C+/ C·C•C·C+ quadruple base pair or C•C·C+ triple base pair stacking on the dimeric block. Our work sheds light on the G-quadruplexes adopted by d(G4C2) and yields the invaluable structural details for the development of small molecules to tackle neurodegenerative diseases, ALS and FTD.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
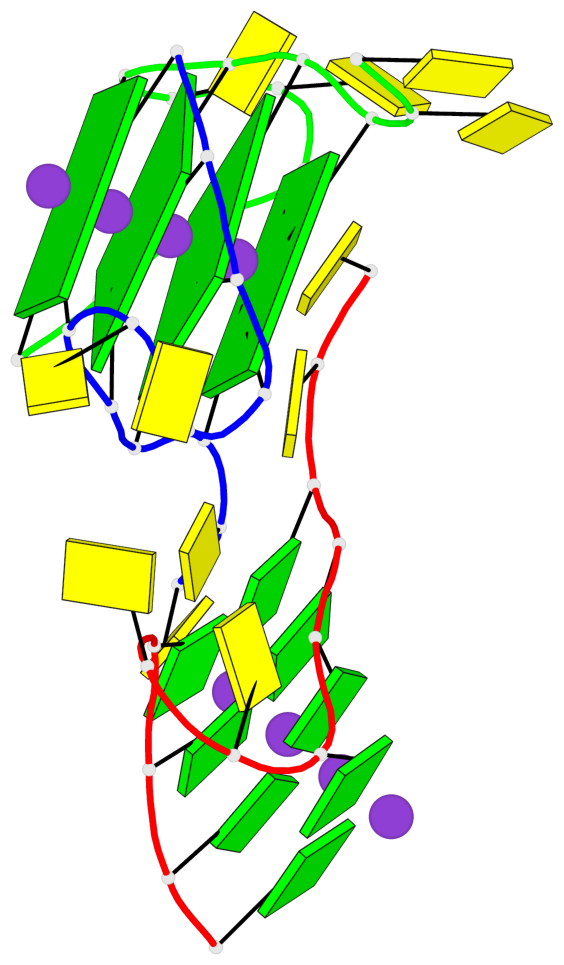
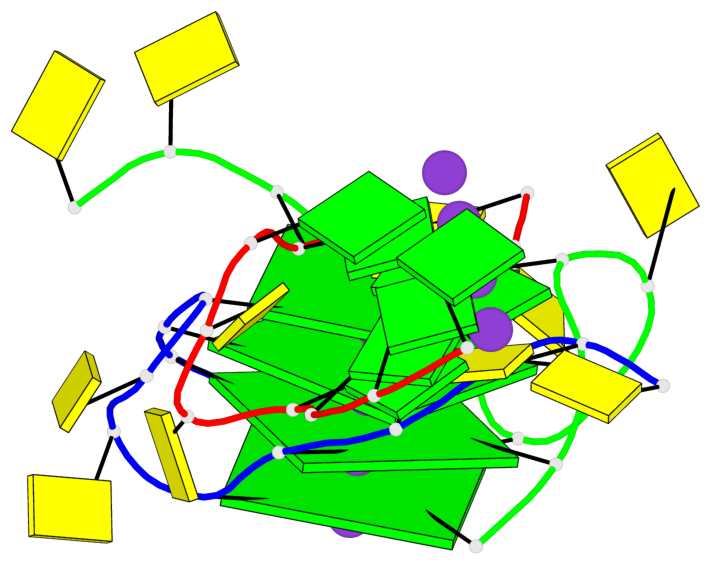
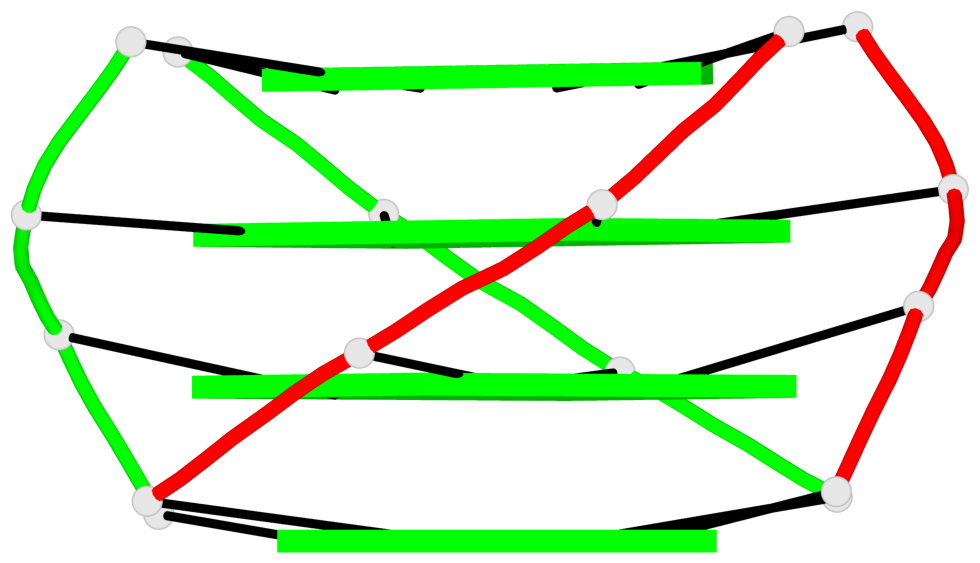
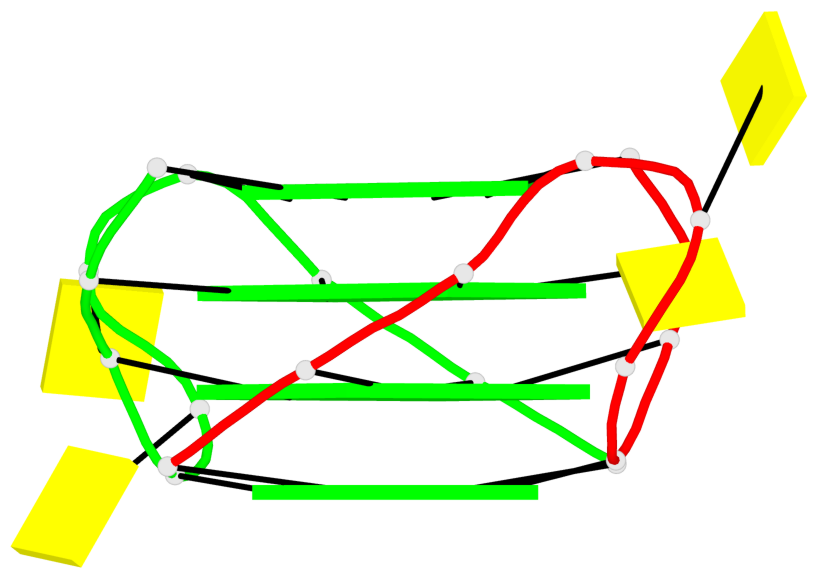
Base-block schematics in six views
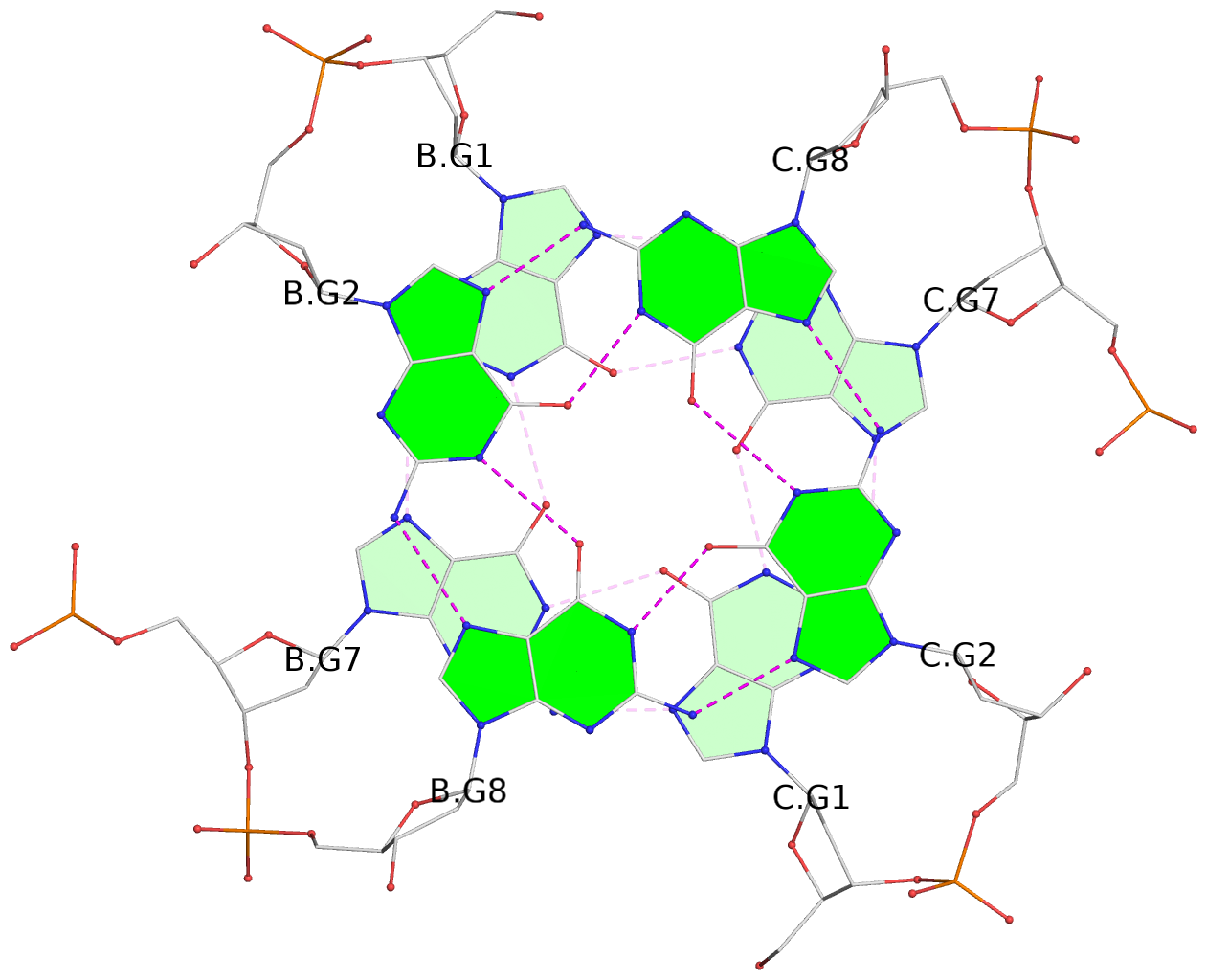
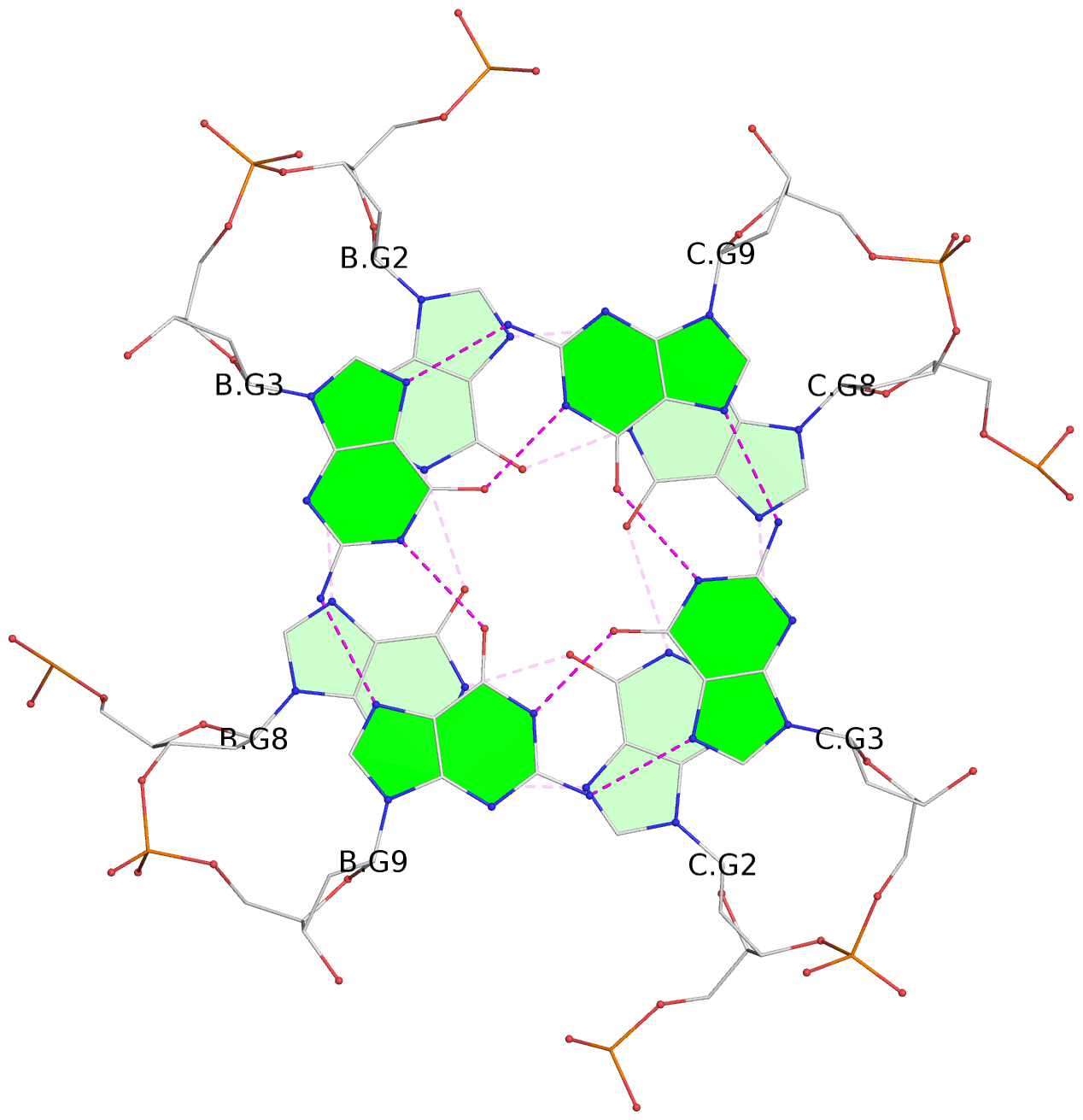
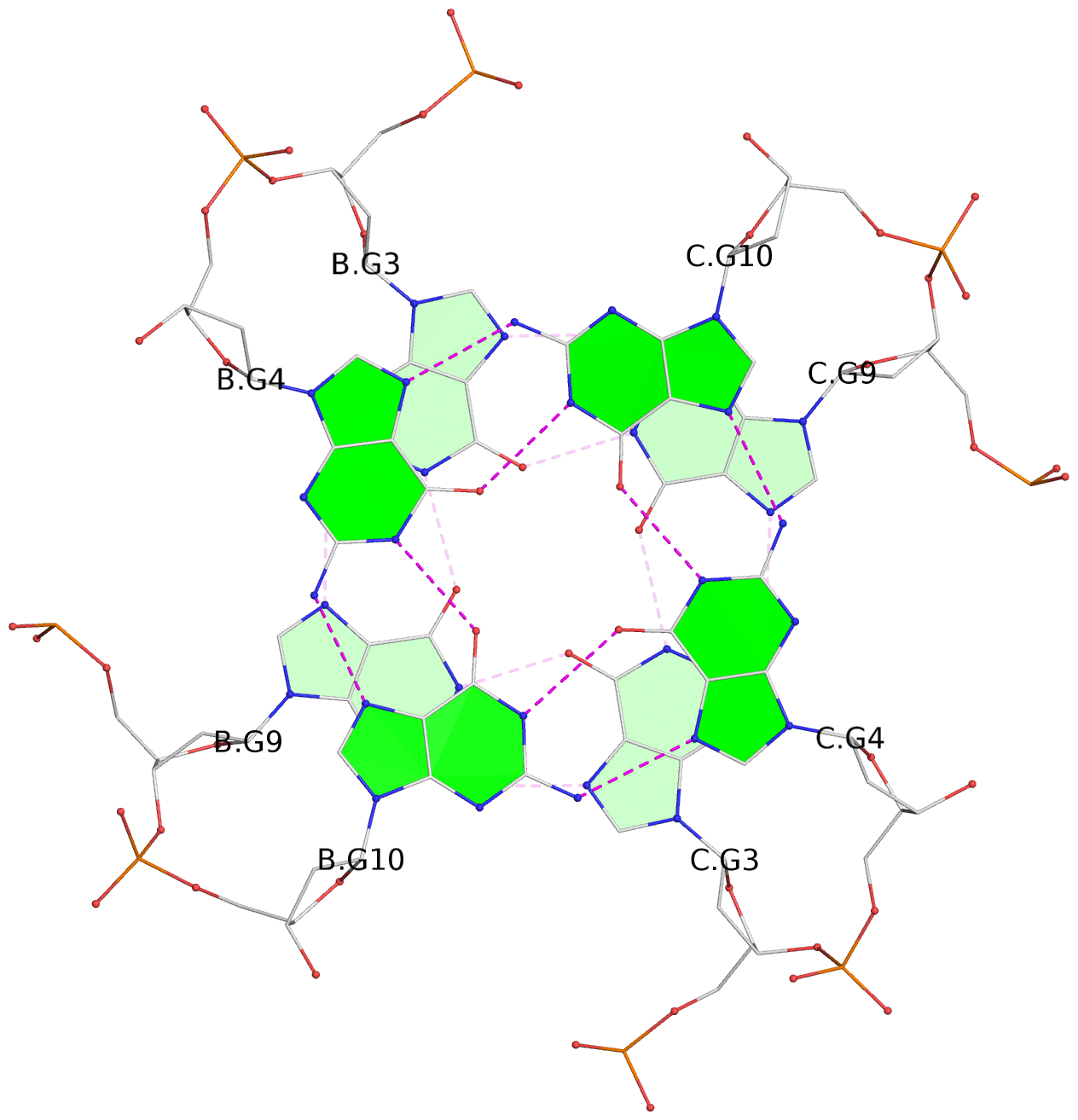
List of 4 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.054 type=planar nts=4 GGGG B.DG1,B.DG7,C.DG1,C.DG7 2 glyco-bond=---- sugar=-.-. groove=---- planarity=0.086 type=planar nts=4 GGGG B.DG2,B.DG8,C.DG2,C.DG8 3 glyco-bond=---- sugar=---- groove=---- planarity=0.120 type=planar nts=4 GGGG B.DG3,B.DG9,C.DG3,C.DG9 4 glyco-bond=---- sugar=---- groove=---- planarity=0.219 type=bowl nts=4 GGGG B.DG4,B.DG10,C.DG4,C.DG10
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.