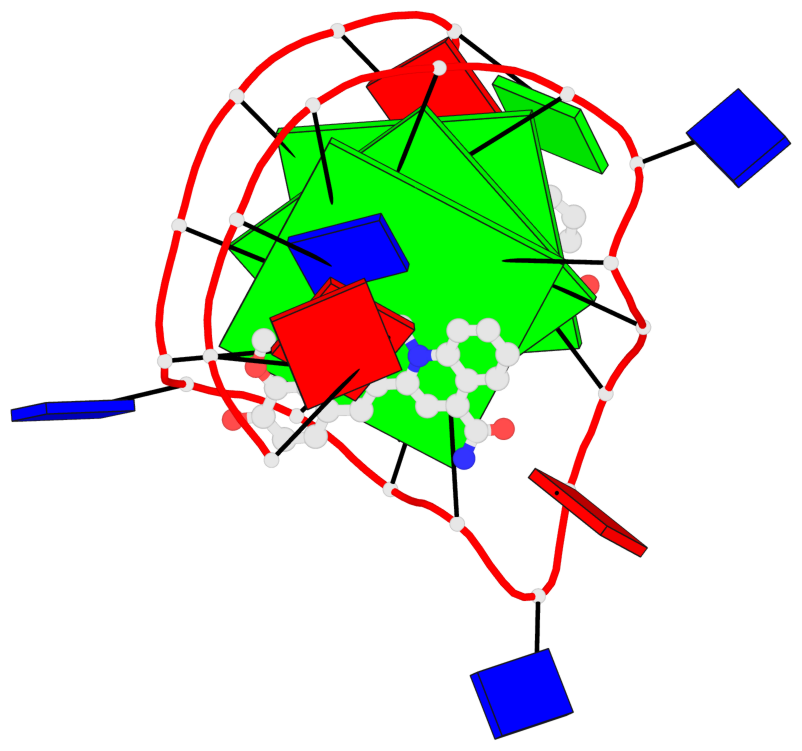
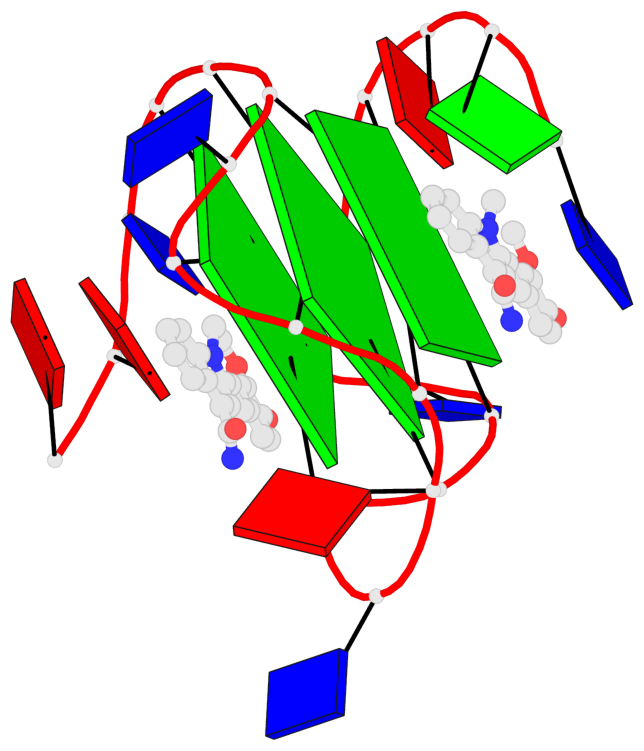
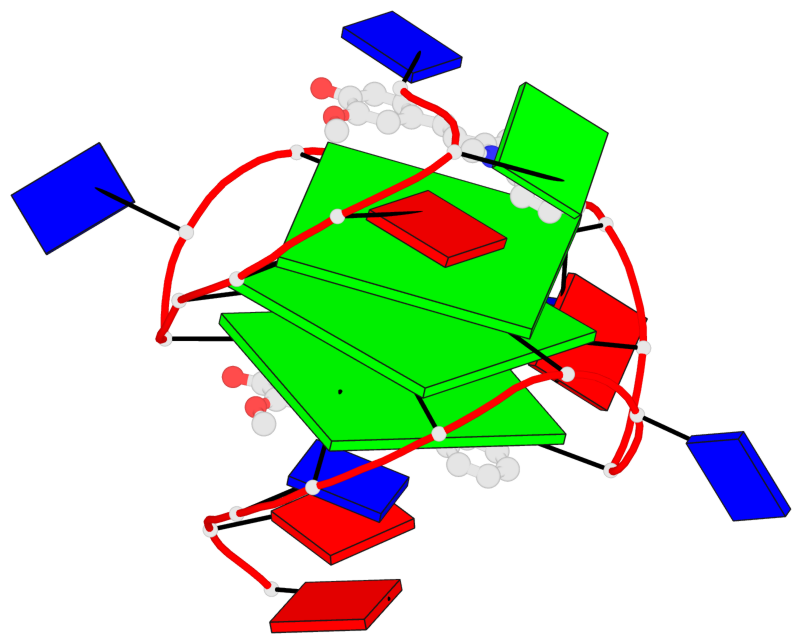
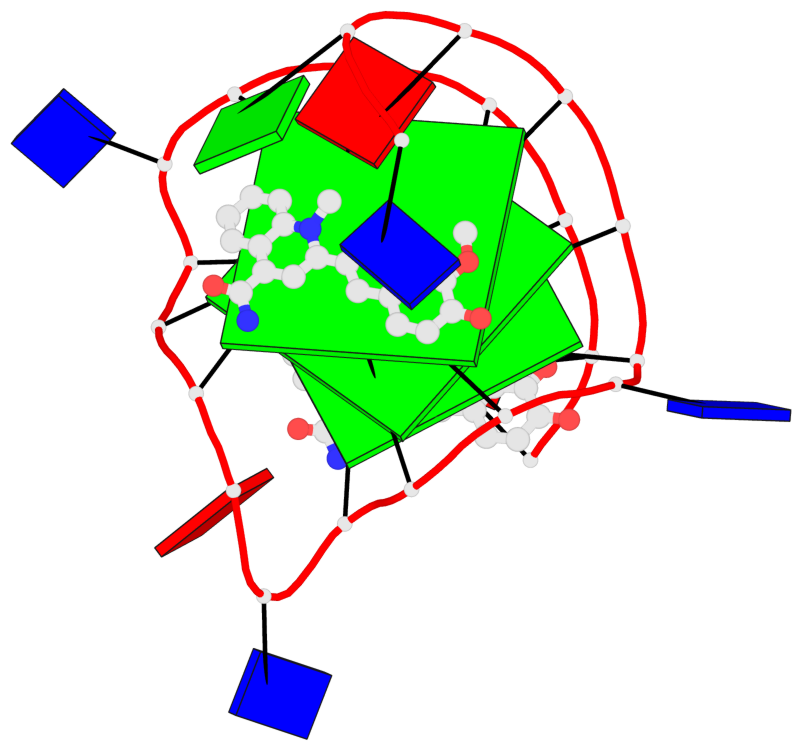
Detailed DSSR results for the G-quadruplex: PDB entry 7kbx
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7kbx
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of the major myc promoter g-quadruplex in complex with nsc85697, a quinoline derivative
- Reference
- Dickerhoff J, Dai J, Yang D (2021): "Structural recognition of the MYC promoter G-quadruplex by a quinoline derivative: insights into molecular targeting of parallel G-quadruplexes." Nucleic Acids Res., 49, 5905-5915. doi: 10.1093/nar/gkab330.
- Abstract
- DNA G-Quadruplexes (G4s) formed in oncogene promoters regulate transcription. The oncogene MYC promoter G4 (MycG4) is the most prevalent G4 in human cancers. However, the most studied MycG4 sequence bears a mutated 3'-residue crucial for ligand recognition. Here, we report a new drug-like small molecule PEQ without a large aromatic moiety that specifically binds MycG4. We determined the NMR solution structures of the wild-type MycG4 and its 2:1 PEQ complex, as well as the structure of the 2:1 PEQ complex of the widely used mutant MycG4. Comparison of the two complex structures demonstrates specific molecular recognition of MycG4 and shows the clear effect of the critical 3'-mutation on the drug binding interface. We performed a systematic analysis of the four available complex structures involving the same mutant MycG4, which can be considered a model system for parallel G4s, and revealed for the first time that the flexible flanking residues are recruited in a conserved and sequence-specific way, as well as unused potential for selective ligand-G4 hydrogen-bond interactions. Our results provide the true molecular basis for MycG4-targeting drugs and new critical insights into future rational design of drugs targeting MycG4 and parallel G4s that are prevalent in promoter and RNA G4s.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
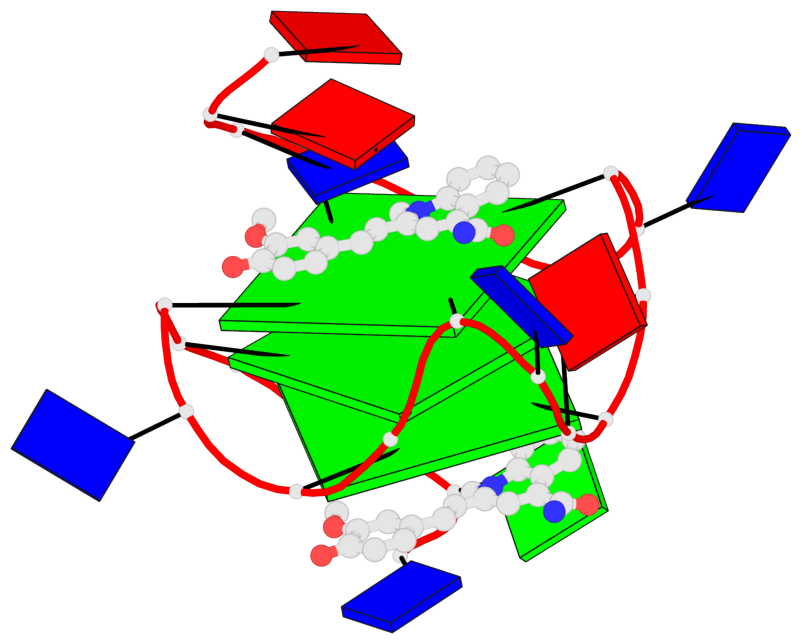
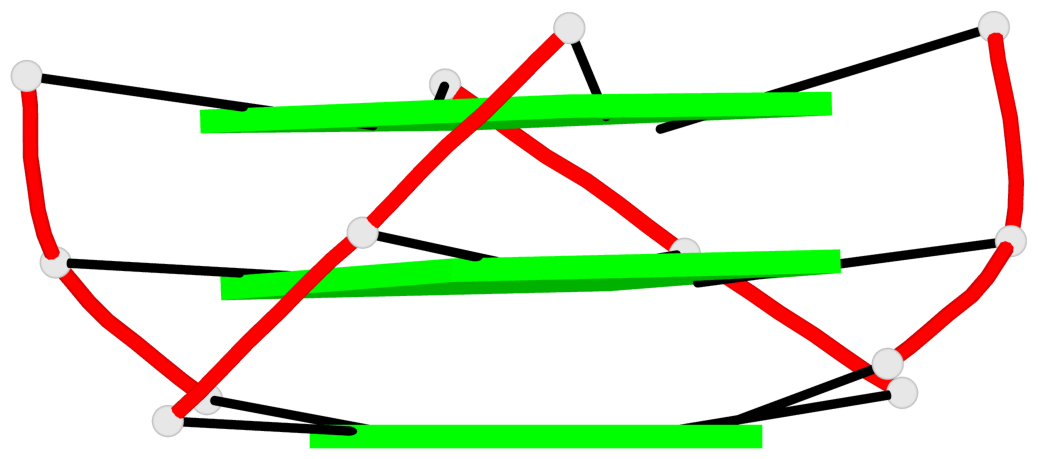
Base-block schematics in six views
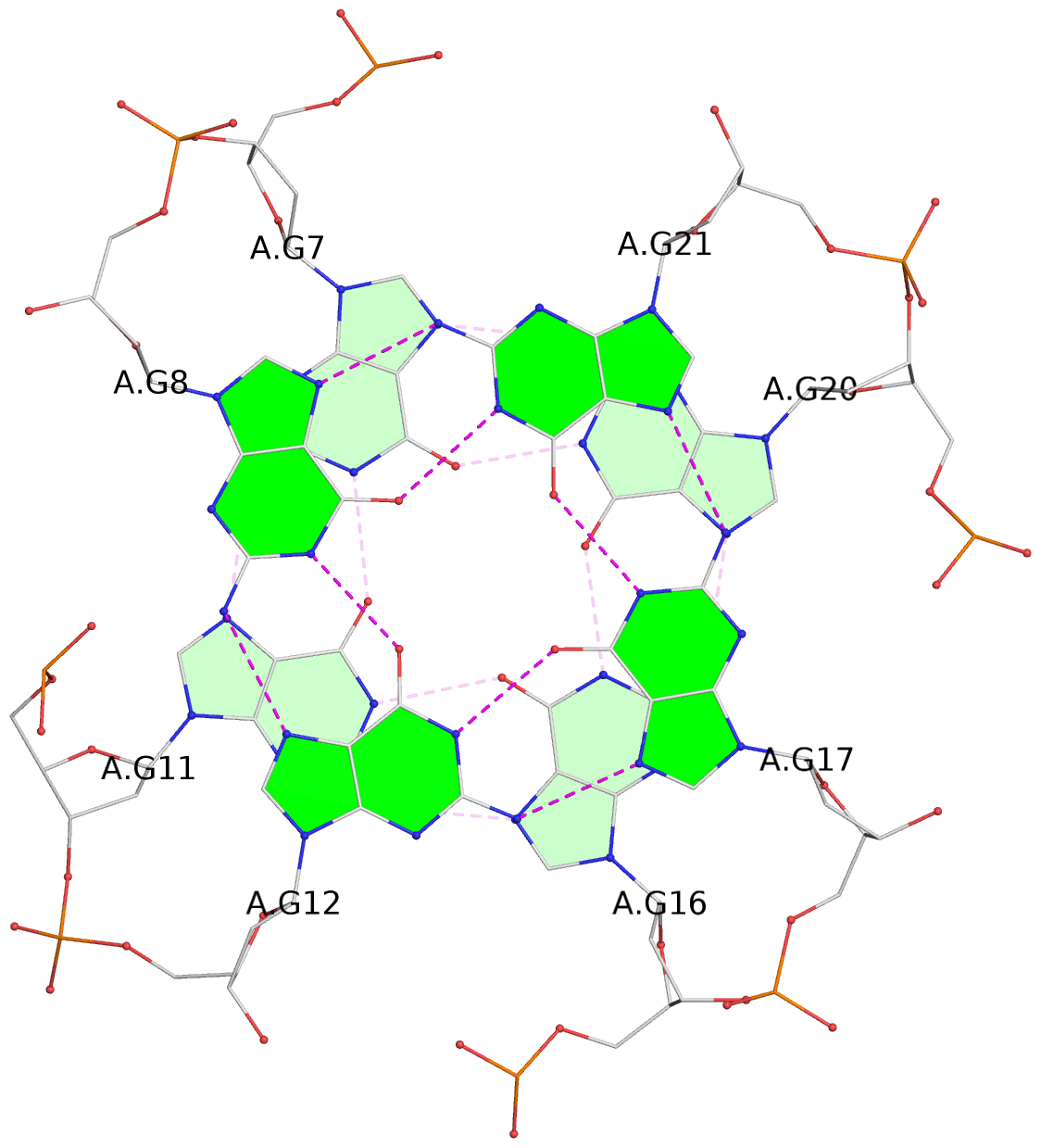
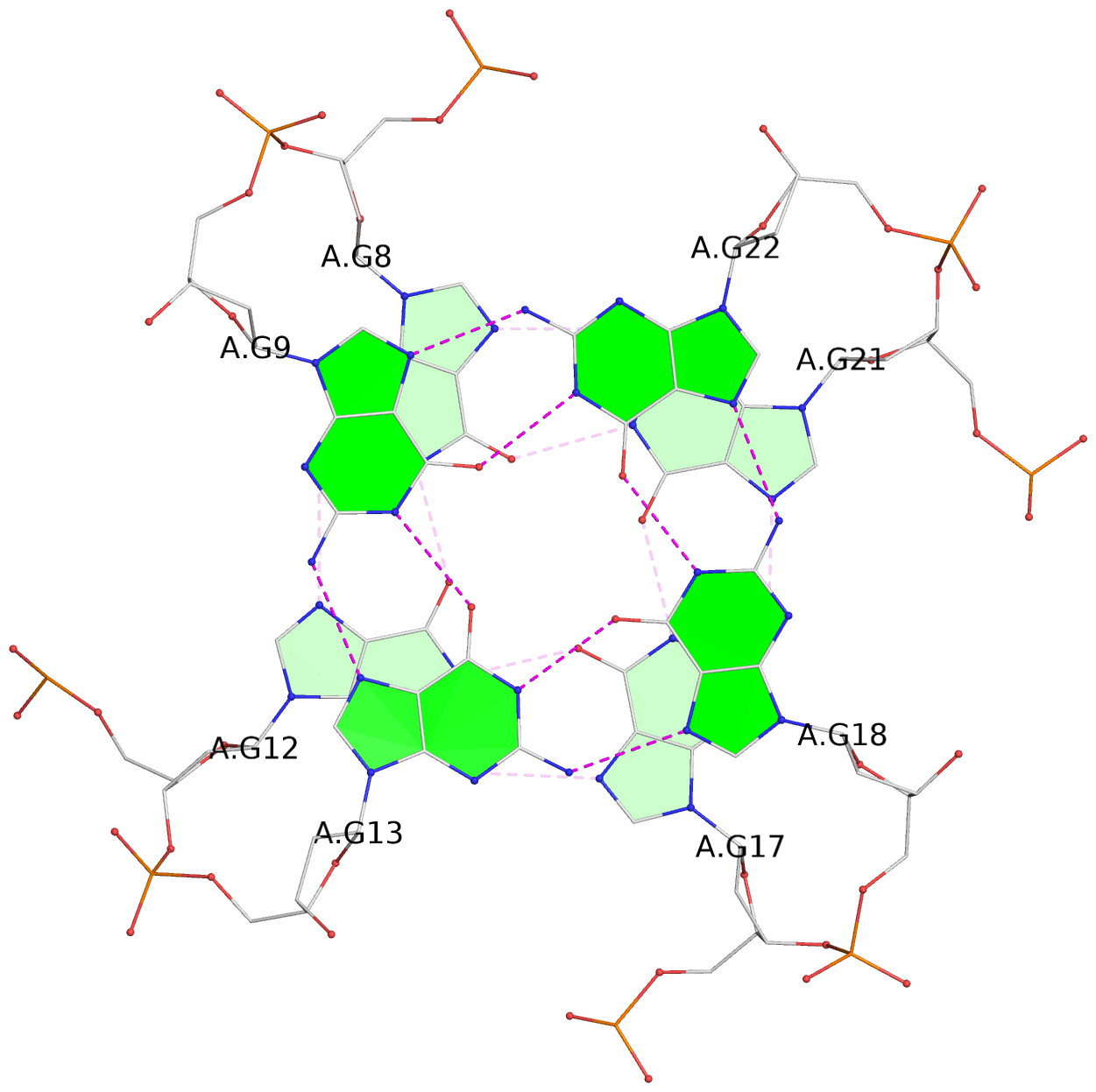
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.157 type=planar nts=4 GGGG A.DG7,A.DG11,A.DG16,A.DG20 2 glyco-bond=---- sugar=---- groove=---- planarity=0.124 type=planar nts=4 GGGG A.DG8,A.DG12,A.DG17,A.DG21 3 glyco-bond=---- sugar=---- groove=---- planarity=0.226 type=bowl nts=4 GGGG A.DG9,A.DG13,A.DG18,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.