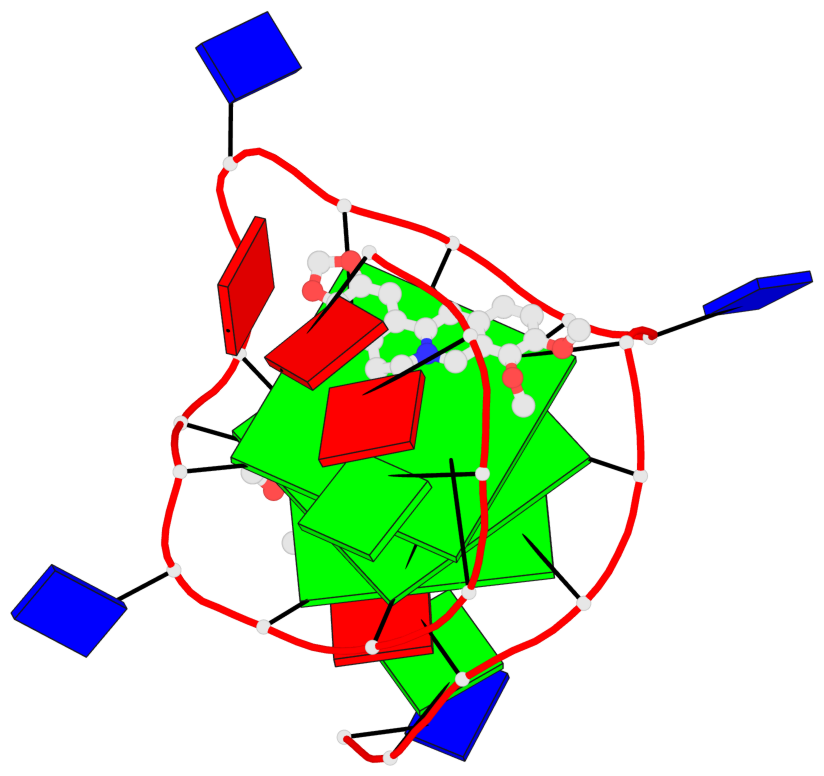
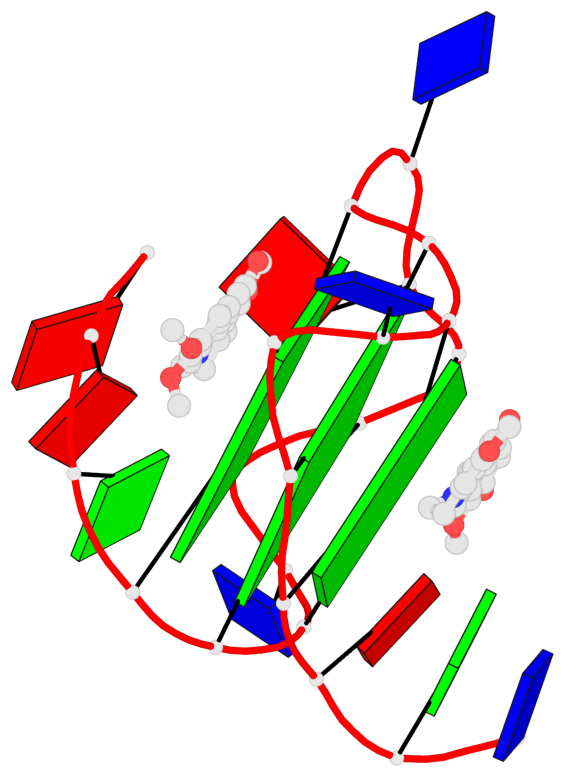
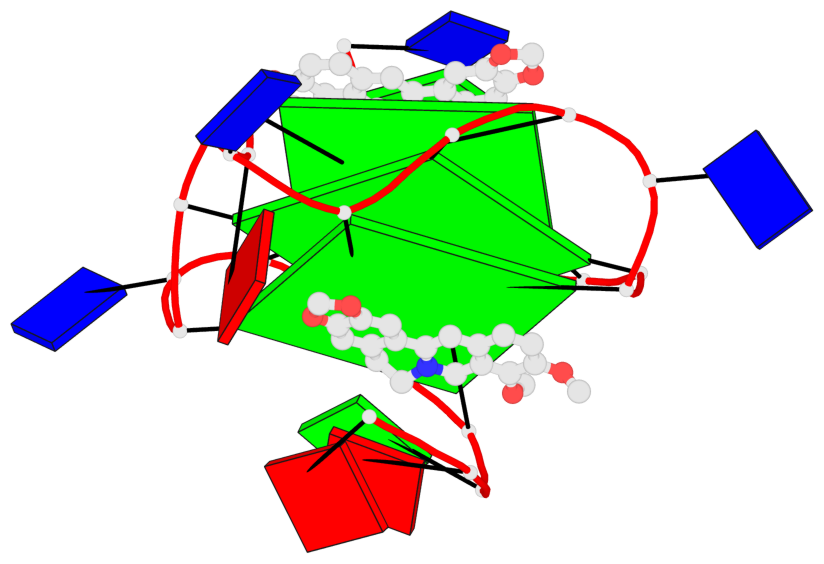
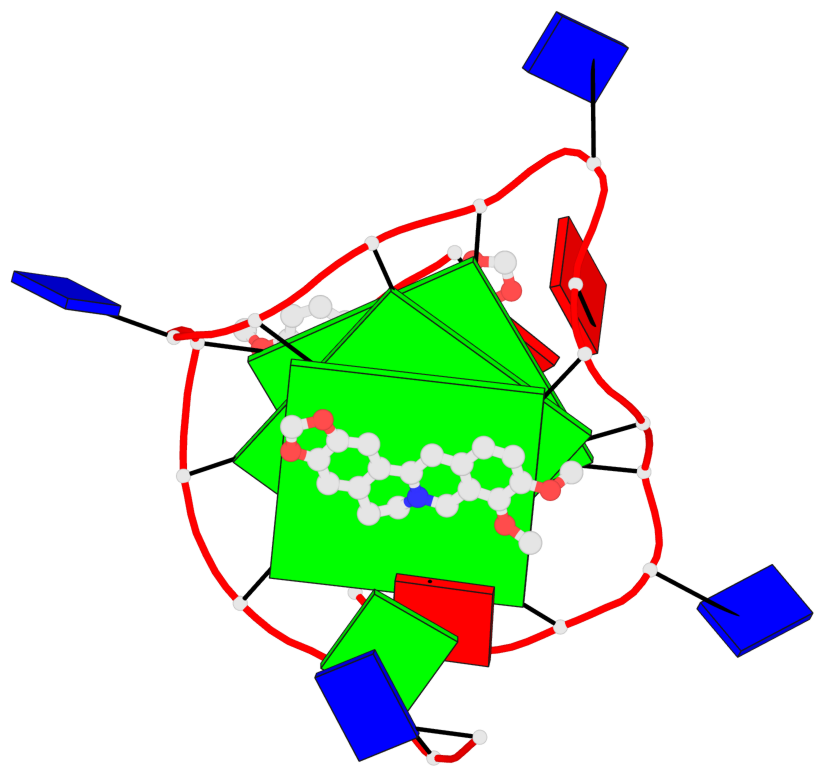
Detailed DSSR results for the G-quadruplex: PDB entry 7n7d
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7n7d
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of the myc promoter g-quadruplex in complex with berberine: conformer a
- Reference
- Dickerhoff J, Brundridge N, McLuckey SA, Yang D (2021): "Berberine Molecular Recognition of the Parallel MYC G-Quadruplex in Solution." J.Med.Chem., 64, 16205-16212. doi: 10.1021/acs.jmedchem.1c01508.
- Abstract
- The medicinal natural product berberine is one of the most actively studied and pursued G-quadruplex (G4)-ligands. The major G-quadruplex formed in the promoter region of the MYC oncogene (MycG4) is an attractive drug target and a prominent example and model structure for parallel G-quadruplexes. G4-targeted berberine derivatives have been actively developed; however, the analogue design was based on a previous crystal structure in which berberine binds as a dimer to a parallel G-quadruplex. Herein, we show that in solution, the binding mode and stoichiometry of berberine are substantially different from the crystal structure: berberine binds as a monomer to MycG4 using a base-recruitment mechanism with a reversed orientation in that the positively charged convex side is actually positioned above the tetrad center. Our structure provides a physiologically relevant basis for the future structure-based rational design of G4-targeted berberine derivatives, and this study demonstrates that it is crucial to validate the ligand-DNA interactions.
- G4 notes
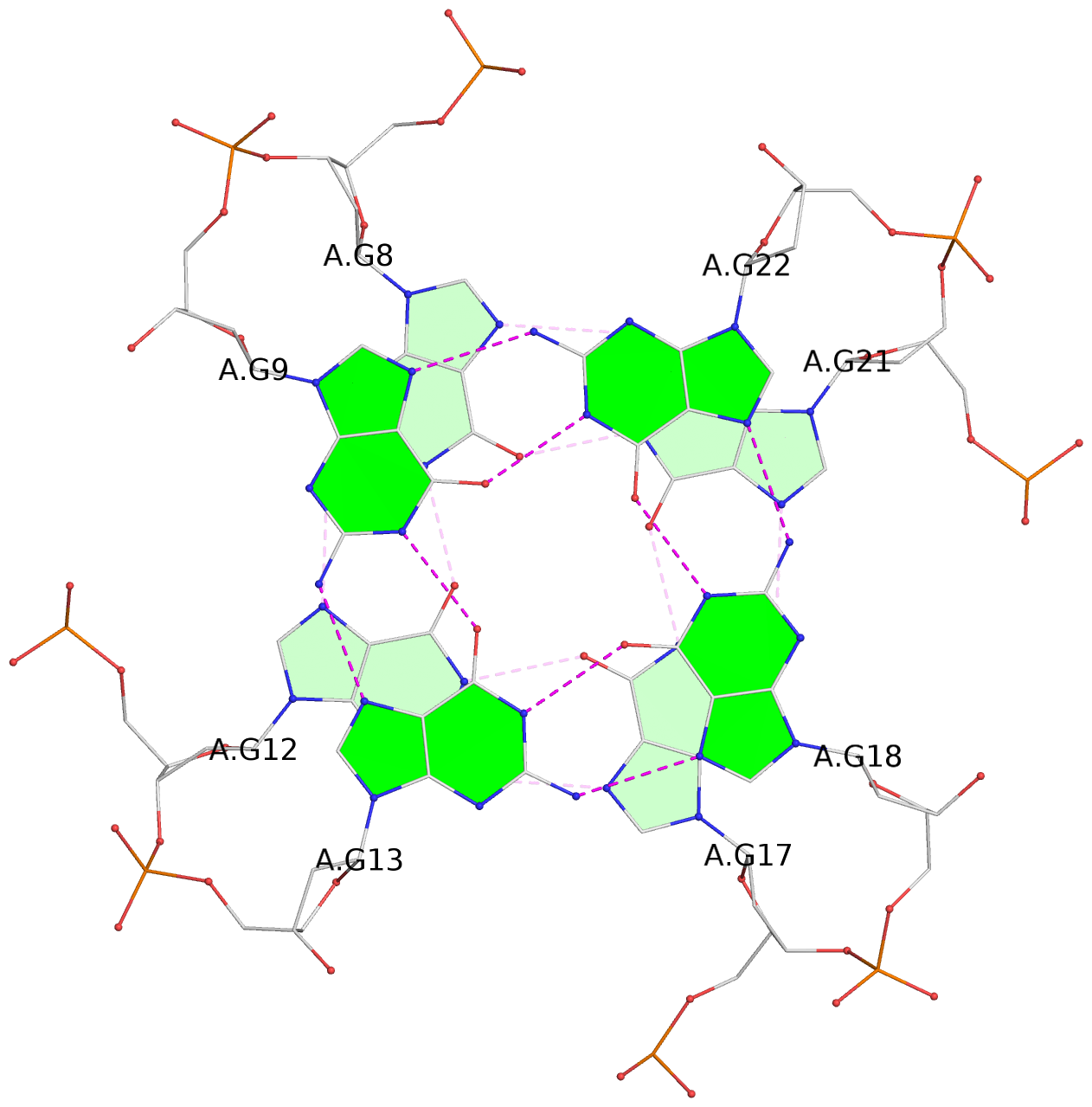
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
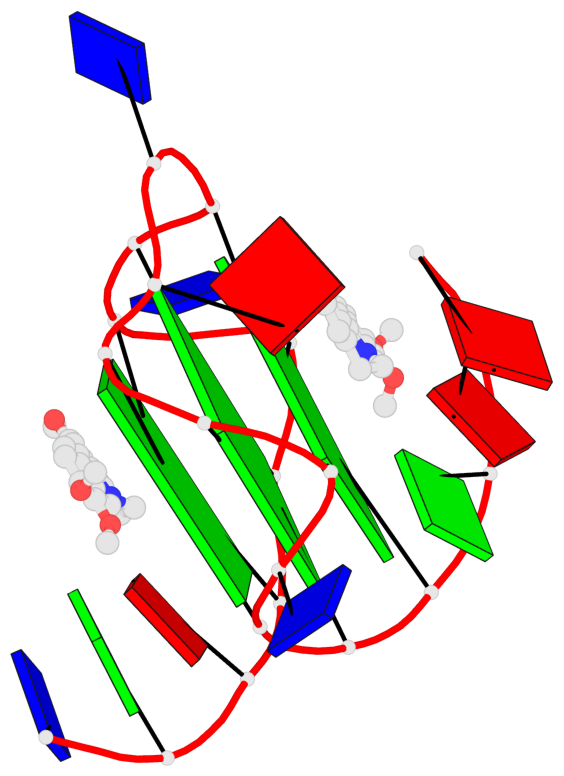
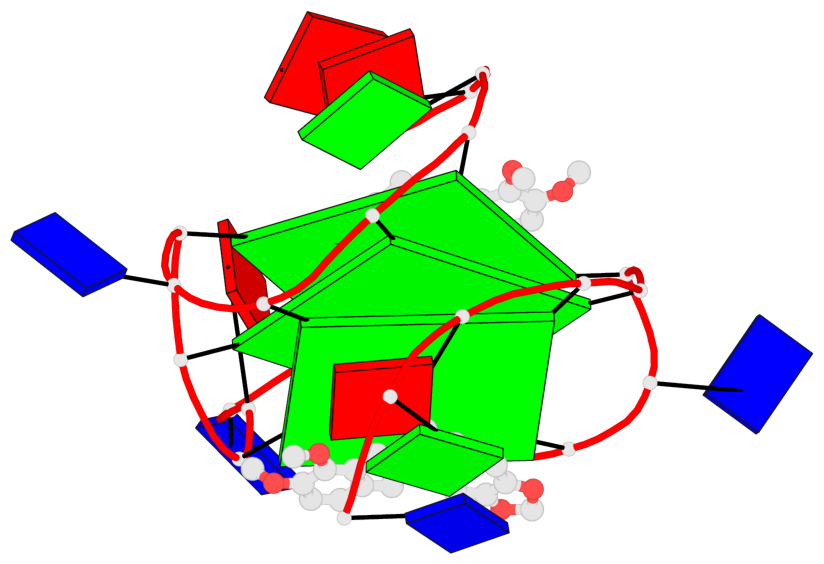
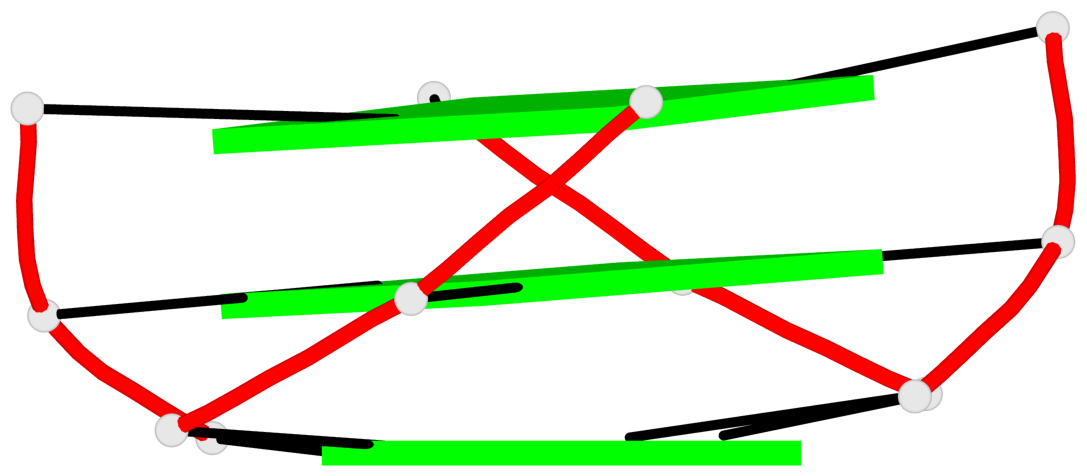
Base-block schematics in six views
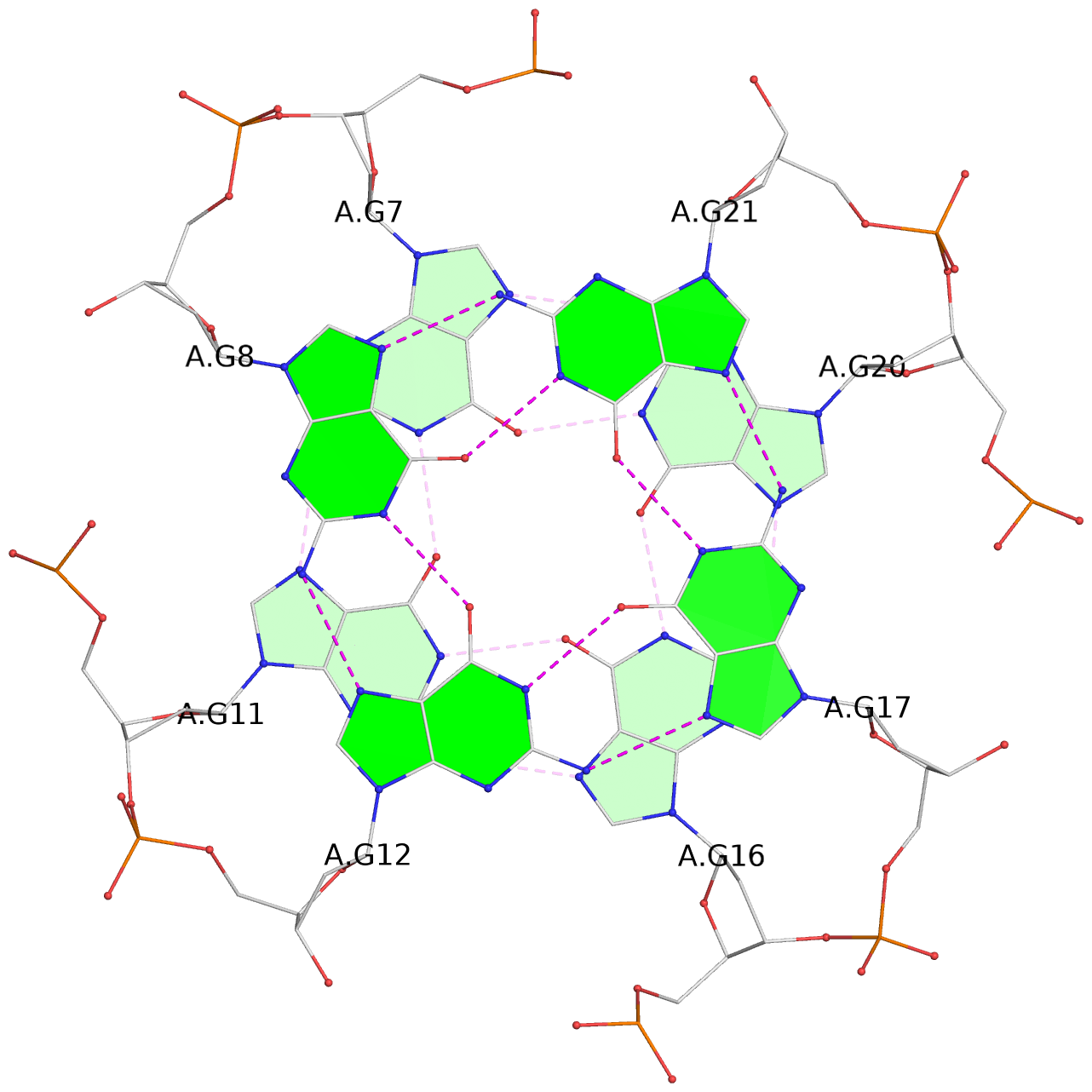
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.197 type=other nts=4 GGGG A.DG7,A.DG11,A.DG16,A.DG20 2 glyco-bond=---- sugar=---- groove=---- planarity=0.157 type=planar nts=4 GGGG A.DG8,A.DG12,A.DG17,A.DG21 3 glyco-bond=---- sugar=---- groove=---- planarity=0.161 type=other nts=4 GGGG A.DG9,A.DG13,A.DG18,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.