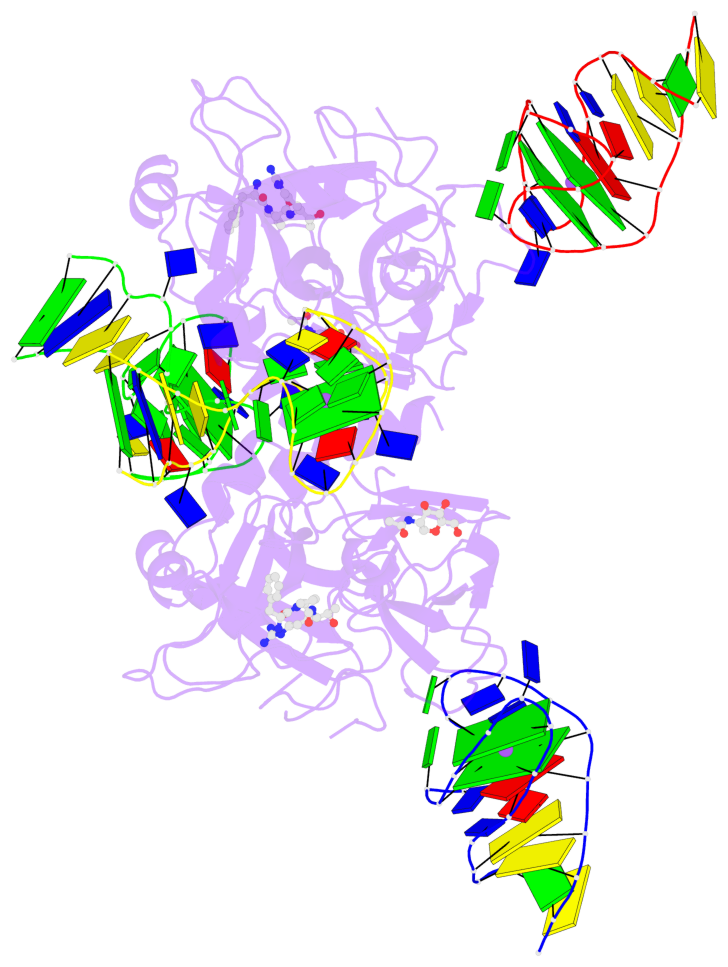
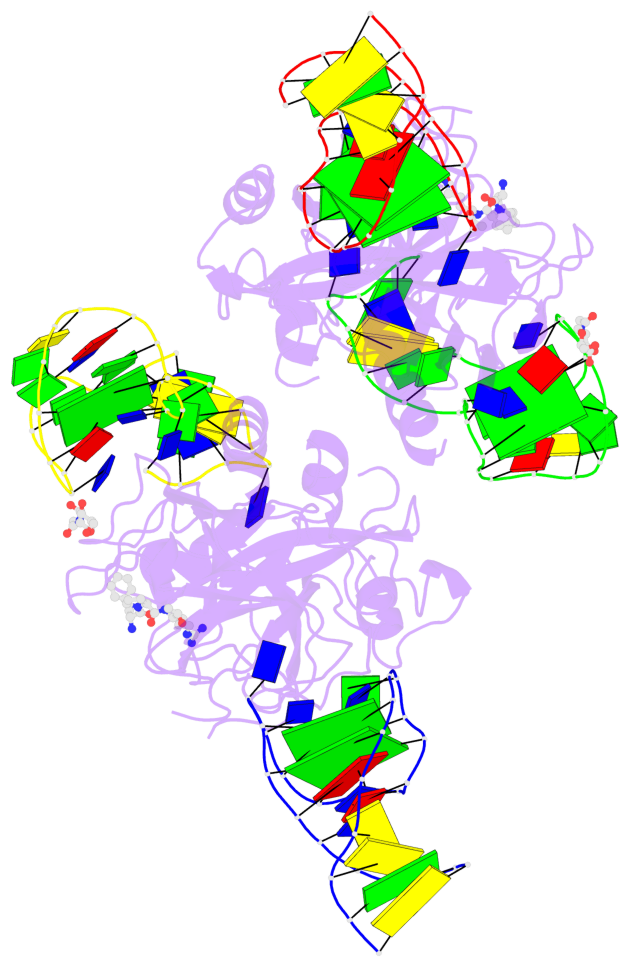
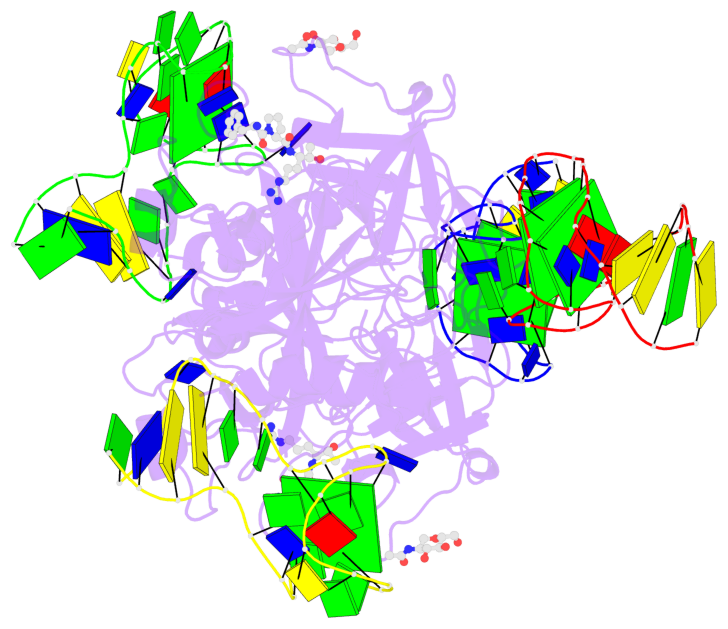
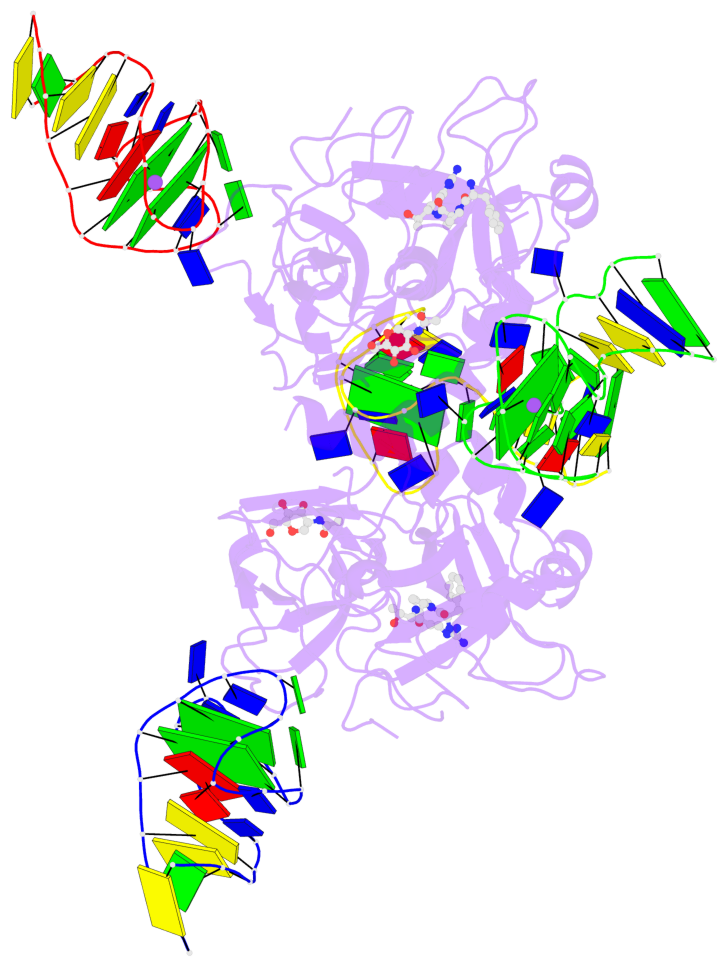
Detailed DSSR results for the G-quadruplex: PDB entry 7ntu
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7ntu
- Class
- hydrolase
- Method
- X-ray (3.1 Å)
- Summary
- X-ray structure of the complex between human alpha thrombin and two duplex-quadruplex aptamers: nu172 and hd22_27mer
- Reference
- Troisi R, Balasco N, Santamaria A, Vitagliano L, Sica F (2021): "Structural and functional analysis of the simultaneous binding of two duplex/quadruplex aptamers to human alpha-thrombin." Int.J.Biol.Macromol., 181, 858-867. doi: 10.1016/j.ijbiomac.2021.04.076.
- Abstract
- The long-range communication between the two exosites of human α-thrombin (thrombin) tightly modulates the protein-effector interactions. Duplex/quadruplex aptamers represent an emerging class of very effective binders of thrombin. Among them, NU172 and HD22 aptamers are at the forefront of exosite I and II recognition, respectively. The present study investigates the simultaneous binding of these two aptamers by combining a structural and dynamics approach. The crystal structure of the ternary complex formed by the thrombin with NU172 and HD22_27mer provides a detailed view of the simultaneous binding of these aptamers to the protein, inspiring the design of novel bivalent thrombin inhibitors. The crystal structure represents the starting model for molecular dynamics study, which points out the cooperation between the binding at the two exosites. In particular, the binding of an aptamer to its exosite reduces the intrinsic flexibility of the other exosite, that preferentially assumes conformations similar to those observed in the bound state, suggesting a predisposition to interact with the other aptamer. This behaviour is reflected in a significant increase of the anticoagulant activity of NU172 when the inactive HD22_27mer is bound to exosite II, providing a clear evidence of the synergic action of the two aptamers.
- G4 notes
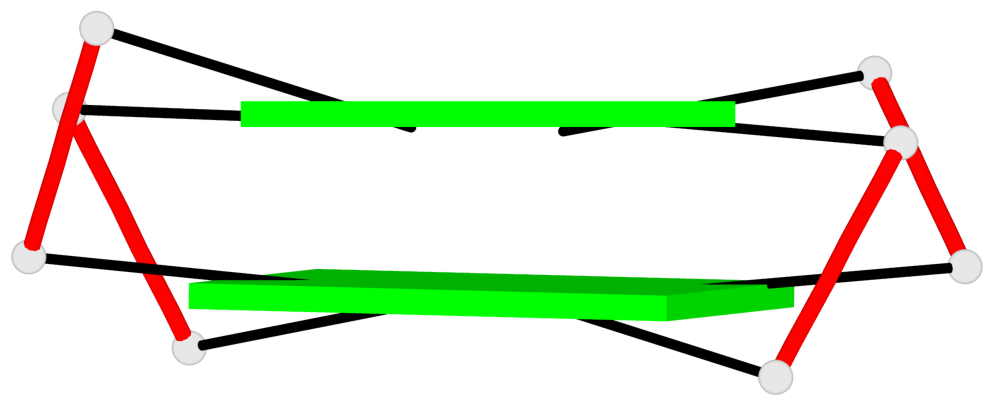
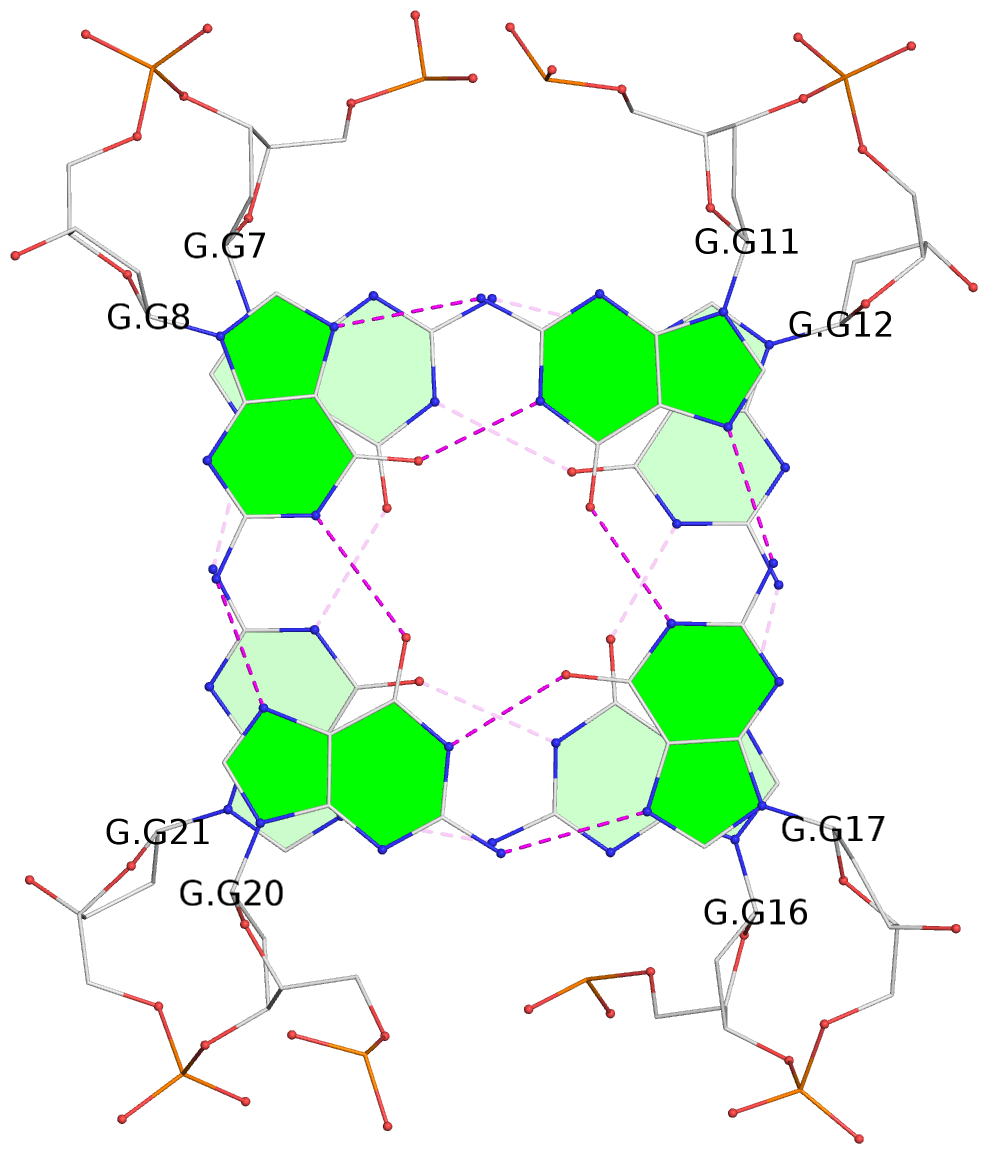
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, 2(+Ln+Lw+Ln), chair(2+2), UDUD
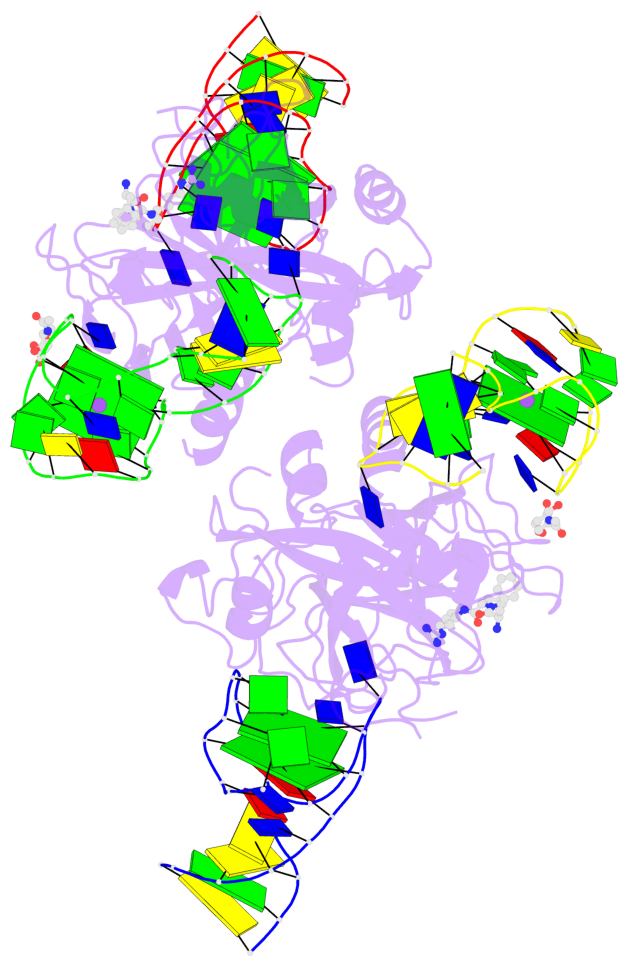
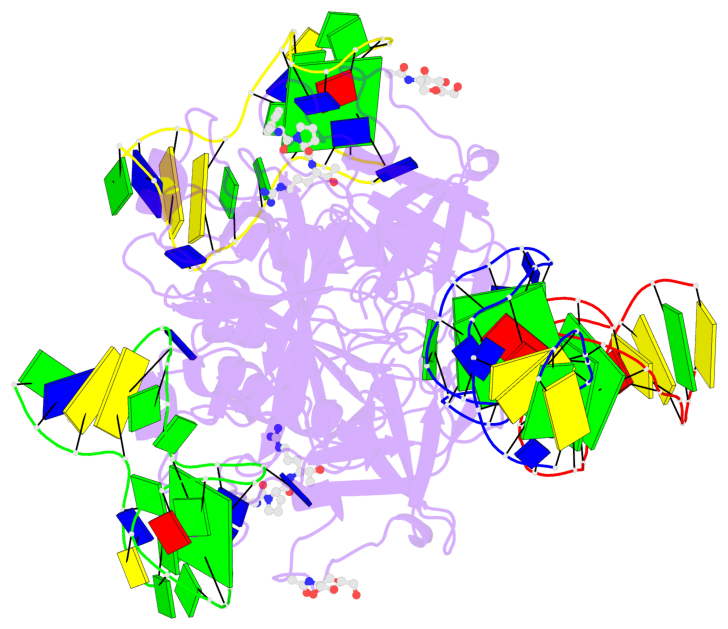
Base-block schematics in six views
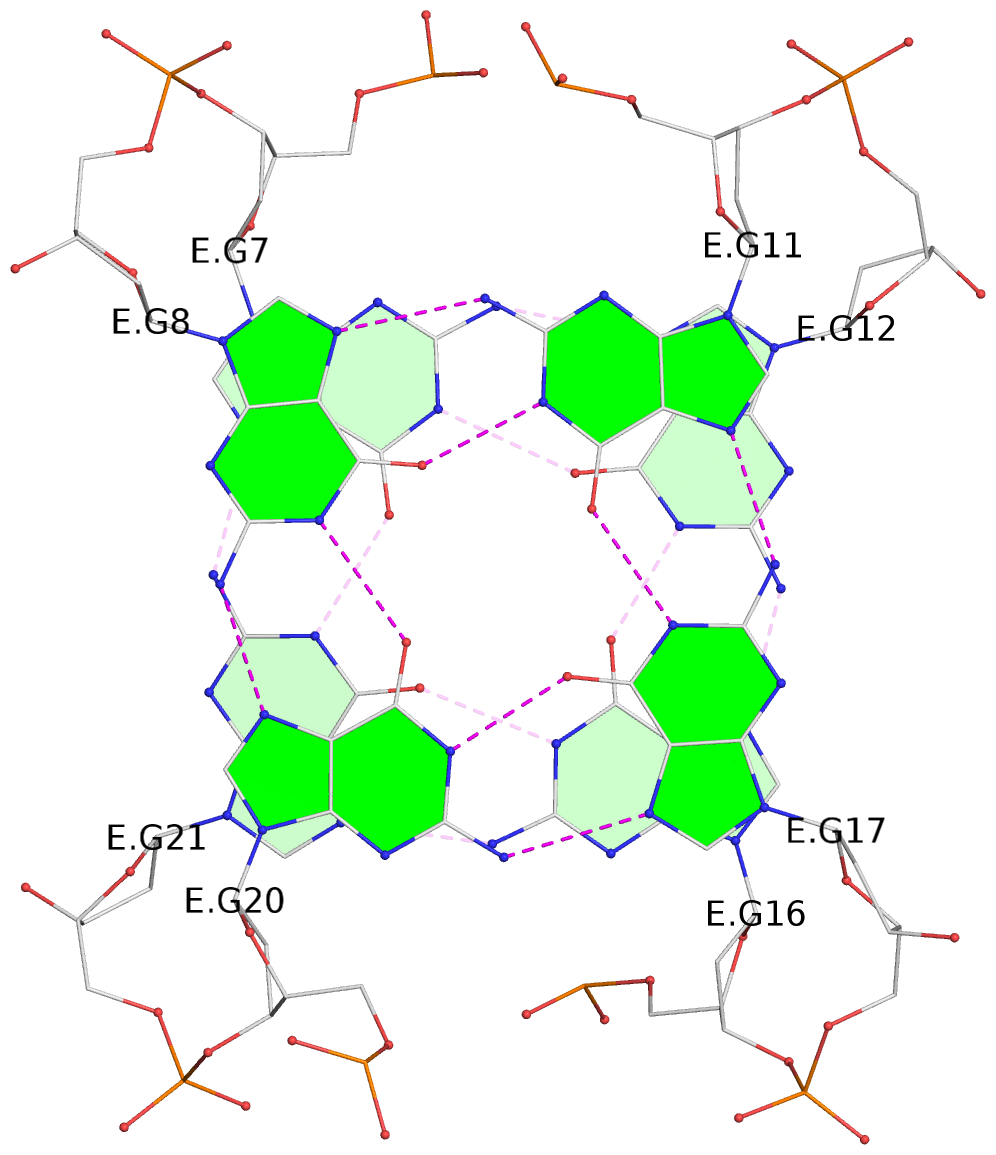
List of 6 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.256 type=other nts=4 GGGG E.DG7,E.DG21,E.DG16,E.DG12 2 glyco-bond=-s-s sugar=--.- groove=wnwn planarity=0.165 type=other nts=4 GGGG E.DG8,E.DG20,E.DG17,E.DG11 3 glyco-bond=-s-s sugar=.3.- groove=wnwn planarity=0.828 type=other nts=4 GGGG F.DG8,F.DG20,F.DG17,F.DG11 4 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.254 type=other nts=4 GGGG G.DG7,G.DG21,G.DG16,G.DG12 5 glyco-bond=-s-s sugar=--.- groove=wnwn planarity=0.169 type=other nts=4 GGGG G.DG8,G.DG20,G.DG17,G.DG11 6 glyco-bond=-s-s sugar=.3.- groove=wnwn planarity=0.841 type=other nts=4 GGGG I.DG8,I.DG20,I.DG17,I.DG11
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDUD, anti-parallel, 2(+Ln+Lw+Ln), chair(2+2)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDUD, anti-parallel, 2(+Ln+Lw+Ln), chair(2+2)
List of 6 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#-1 nts=4 GTAG F.DG8,F.DT9,F.DA10,F.DG11 2 type=lateral helix=#-1 nts=7 GGGCAGG F.DG11,F.DG12,F.DG13,F.DC14,F.DA15,F.DG16,F.DG17 3 type=lateral helix=#-1 nts=4 GTTG F.DG17,F.DT18,F.DT19,F.DG20 4 type=lateral helix=#-2 nts=4 GTAG I.DG8,I.DT9,I.DA10,I.DG11 5 type=lateral helix=#-2 nts=7 GGGCAGG I.DG11,I.DG12,I.DG13,I.DC14,I.DA15,I.DG16,I.DG17 6 type=lateral helix=#-2 nts=4 GTTG I.DG17,I.DT18,I.DT19,I.DG20