Detailed DSSR results for the G-quadruplex: PDB entry 7nwd
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7nwd
- Class
- DNA
- Method
- NMR
- Summary
- Three-quartet c-kit2 g-quadruplex stabilized by a pyrene conjugate
- Reference
- Peterkova K, Durnik I, Marek R, Plavec J, Podbevsek P (2021): "c-kit2 G-quadruplex stabilized via a covalent probe: exploring G-quartet asymmetry." Nucleic Acids Res., 49, 8947-8960. doi: 10.1093/nar/gkab659.
- Abstract
- Several sequences forming G-quadruplex are highly conserved in regulatory regions of genomes of different organisms and affect various biological processes like gene expression. Diverse G-quadruplex properties can be modulated via their interaction with small polyaromatic molecules such as pyrene. To investigate how pyrene interacts with G-rich DNAs, we incorporated deoxyuridine nucleotide(s) with a covalently attached pyrene moiety (Upy) into a model system that forms parallel G-quadruplex structures. We individually substituted terminal positions and positions in the pentaloop of the c-kit2 sequence originating from the KIT proto-oncogene with Upy and performed a detailed NMR structural study accompanied with molecular dynamic simulations. Our results showed that incorporation into the pentaloop leads to structural polymorphism and in some cases also thermal destabilization. In contrast, terminal positions were found to cause a substantial thermodynamic stabilization while preserving topology of the parent c-kit2 G-quadruplex. Thermodynamic stabilization results from π-π stacking between the polyaromatic core of the pyrene moiety and guanine nucleotides of outer G-quartets. Thanks to the prevalent overall conformation, our structures mimic the G-quadruplex found in human KIT proto-oncogene and could potentially have antiproliferative effects on cancer cells.
- G4 notes
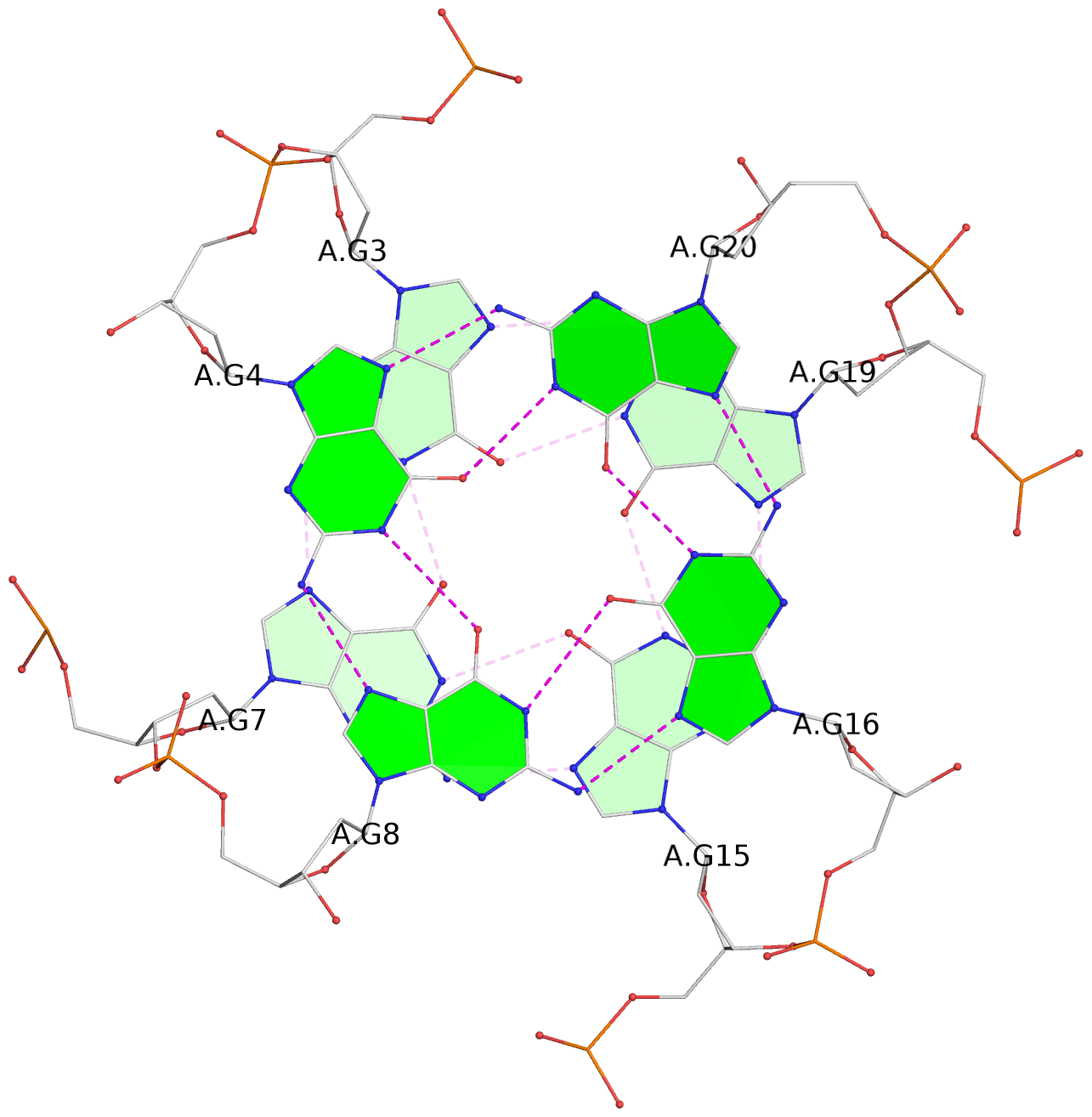
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
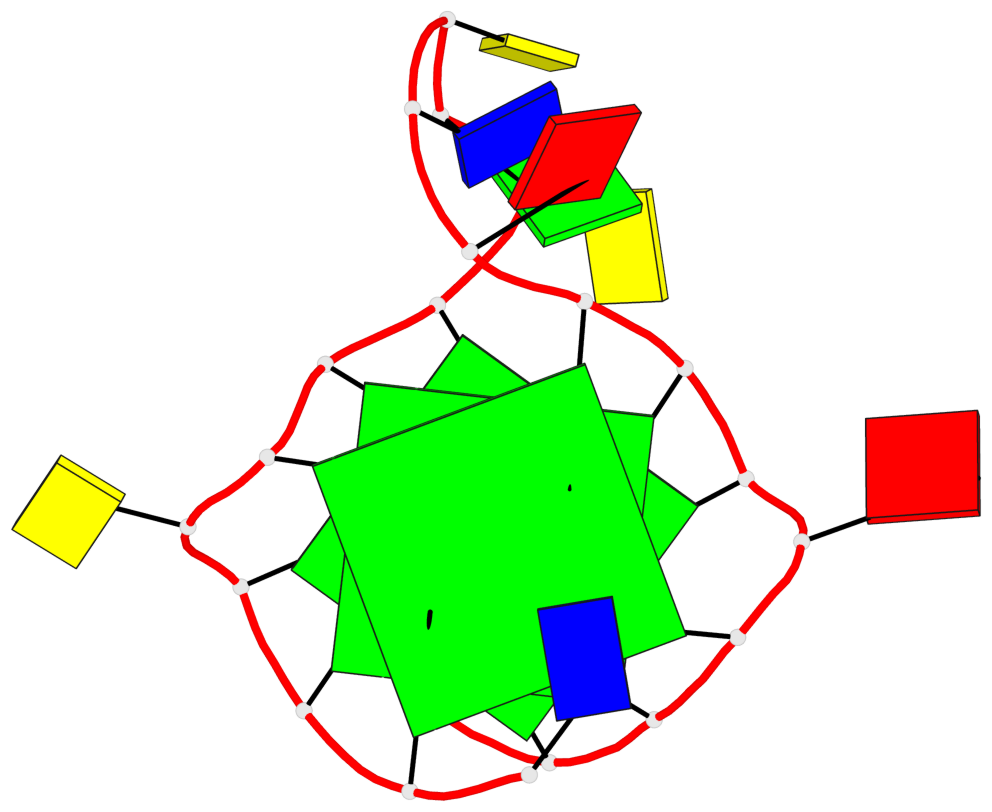
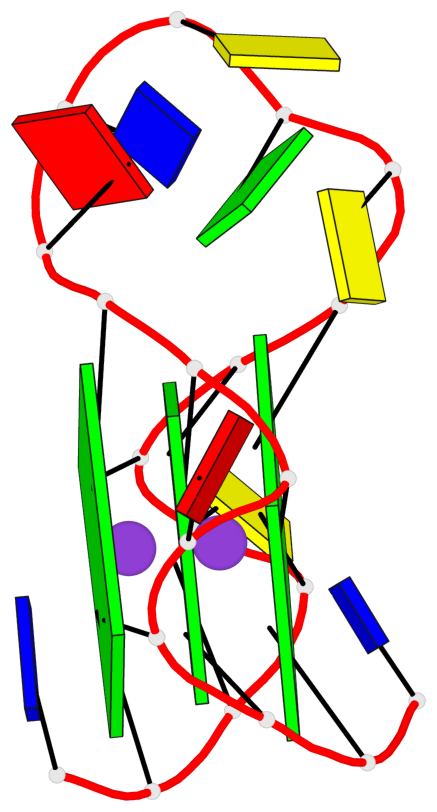
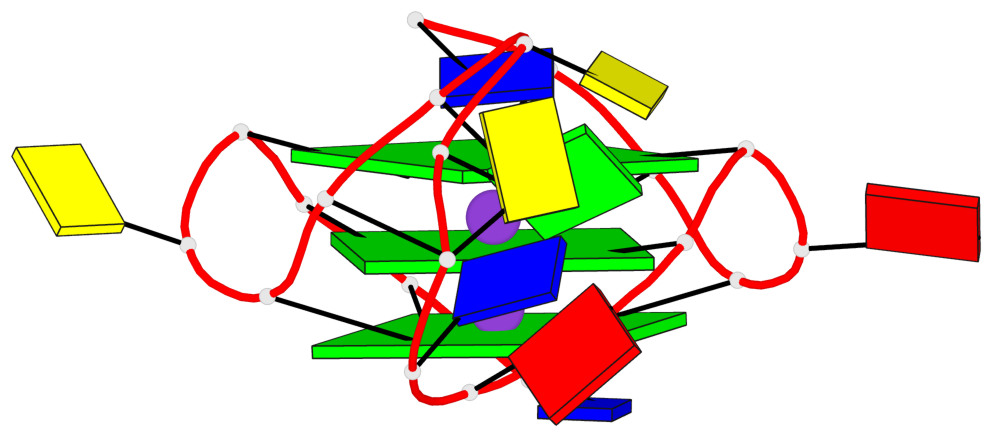
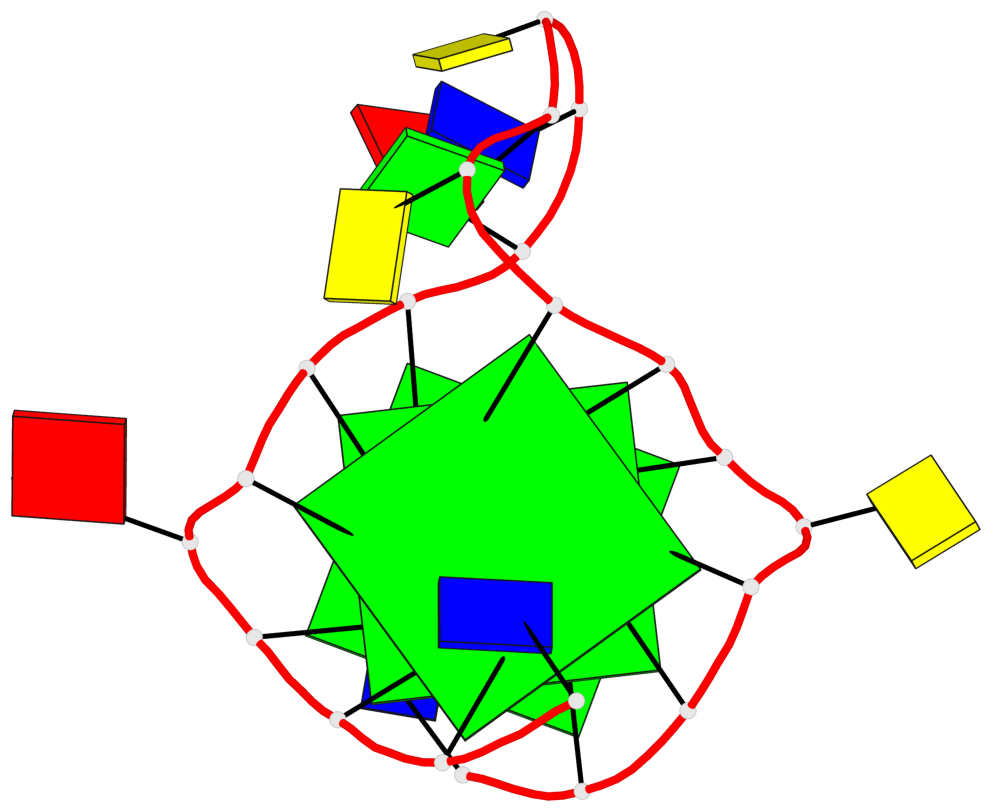
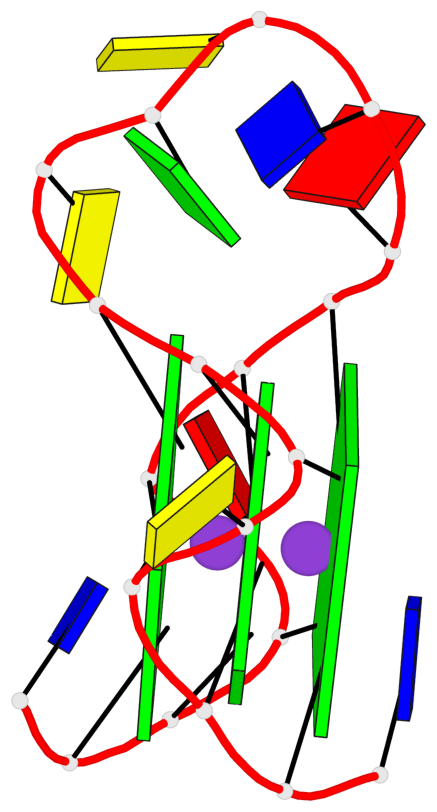
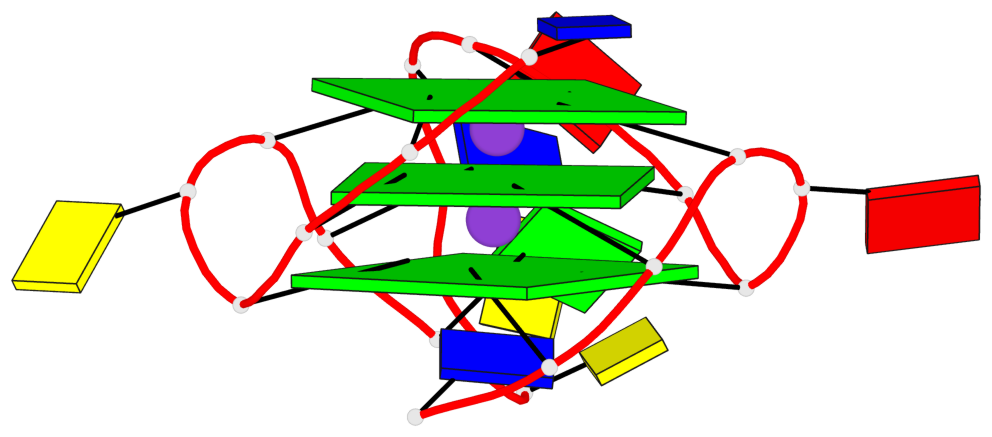
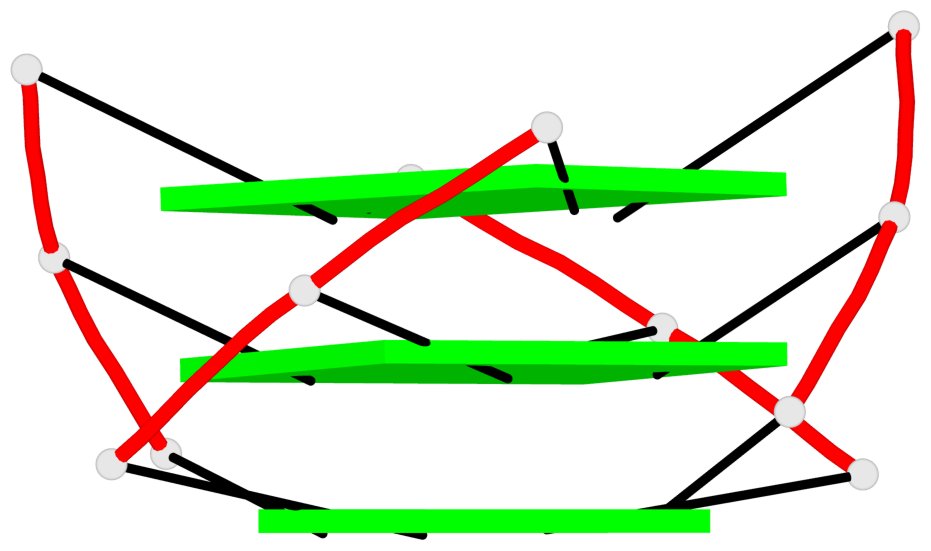
Base-block schematics in six views
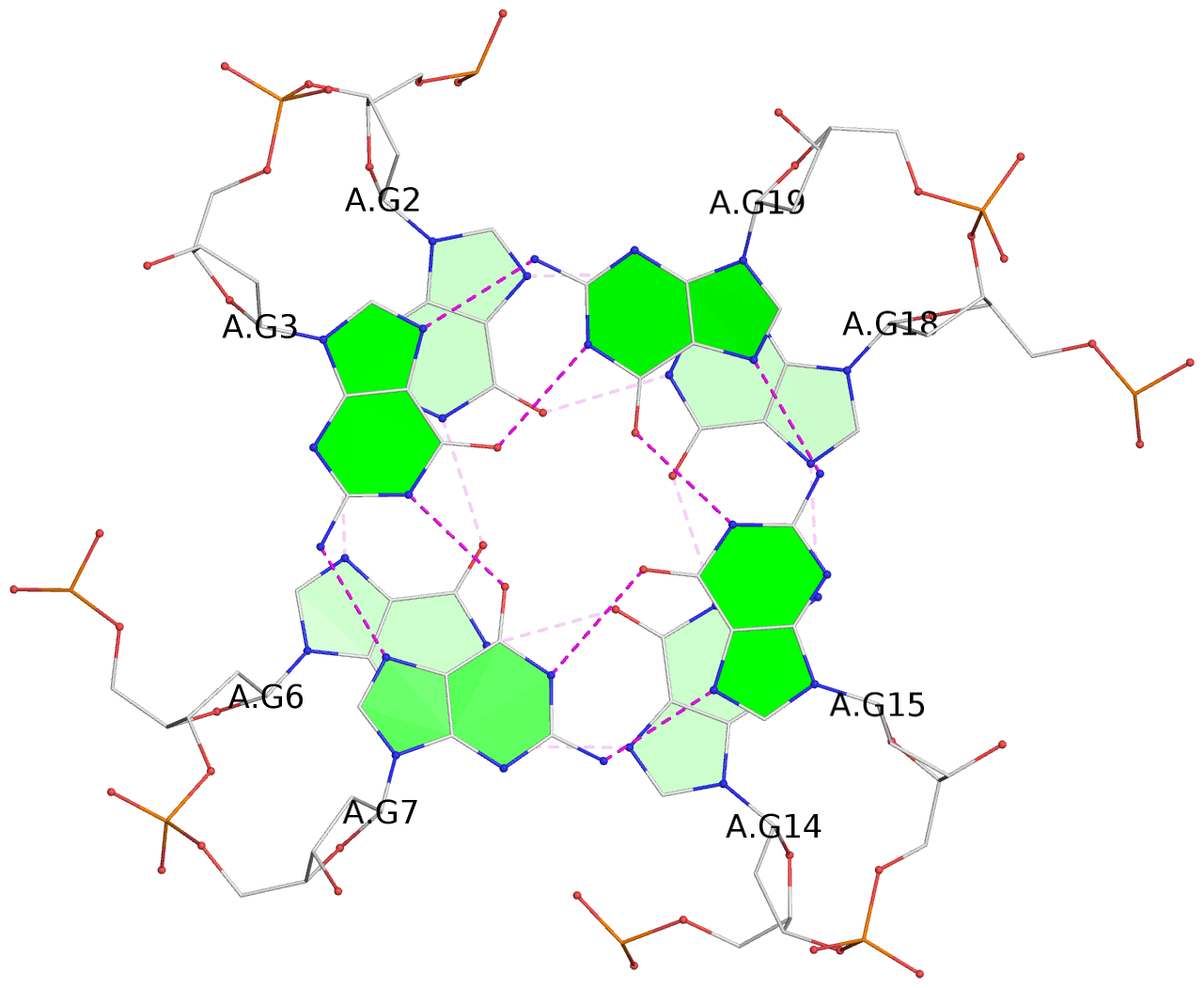
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.240 type=bowl nts=4 GGGG A.DG2,A.DG6,A.DG14,A.DG18 2 glyco-bond=---- sugar=---- groove=---- planarity=0.349 type=bowl nts=4 GGGG A.DG3,A.DG7,A.DG15,A.DG19 3 glyco-bond=---- sugar=---- groove=---- planarity=0.503 type=bowl nts=4 GGGG A.DG4,A.DG8,A.DG16,A.DG20
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.