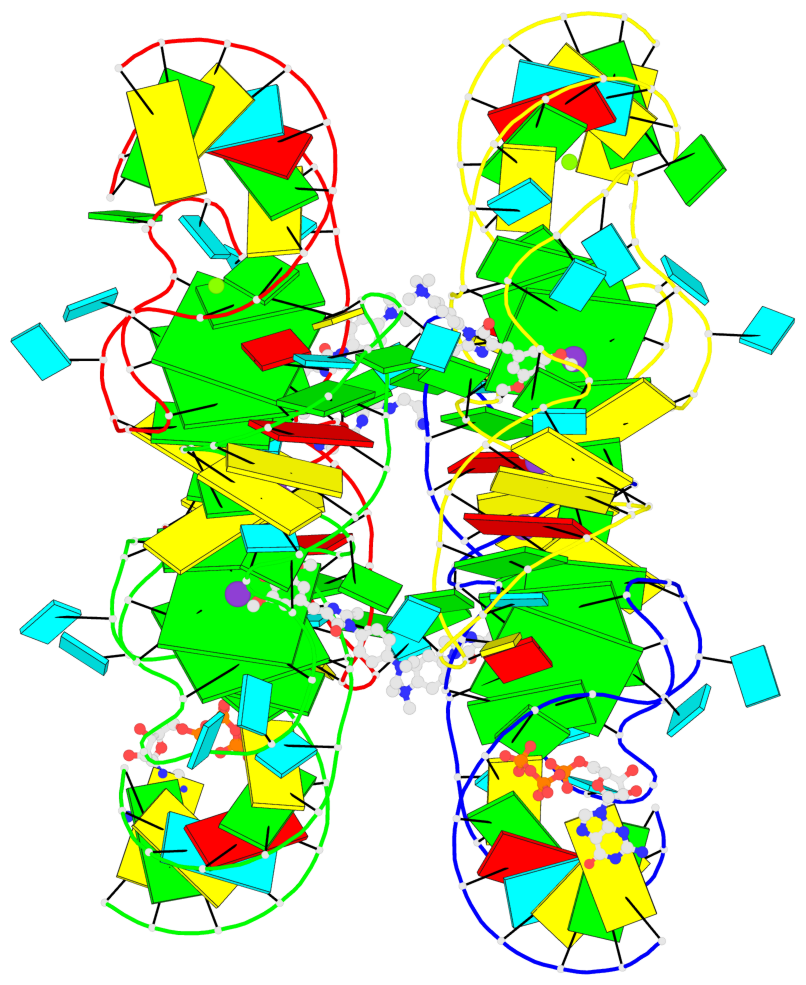
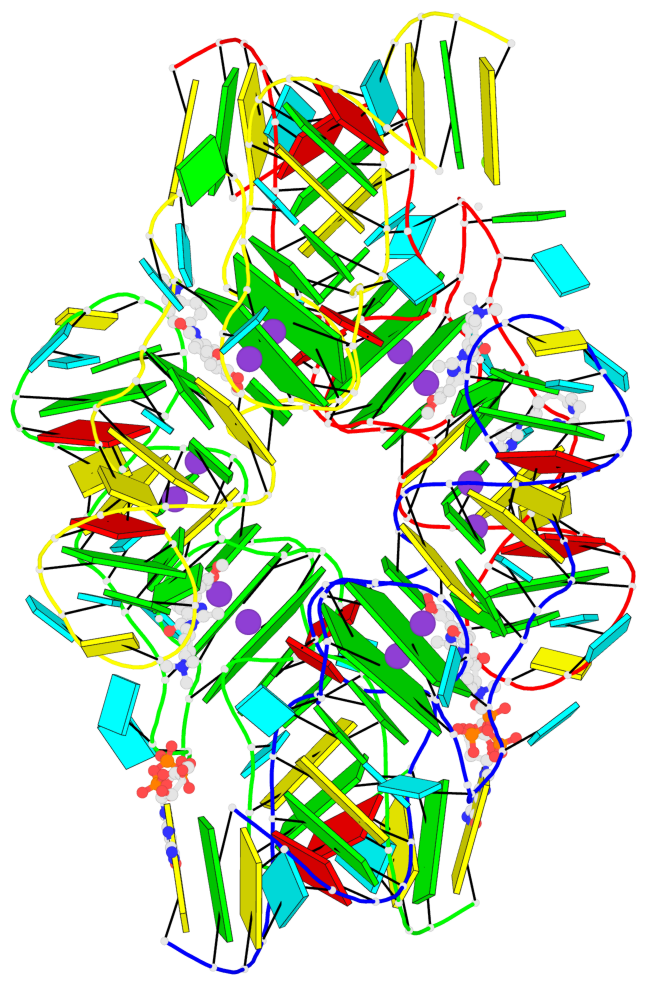
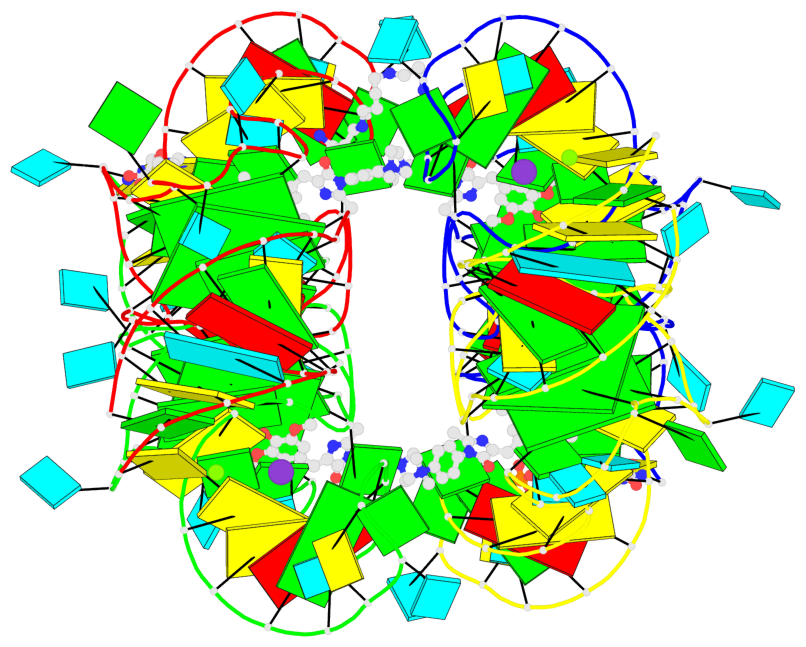
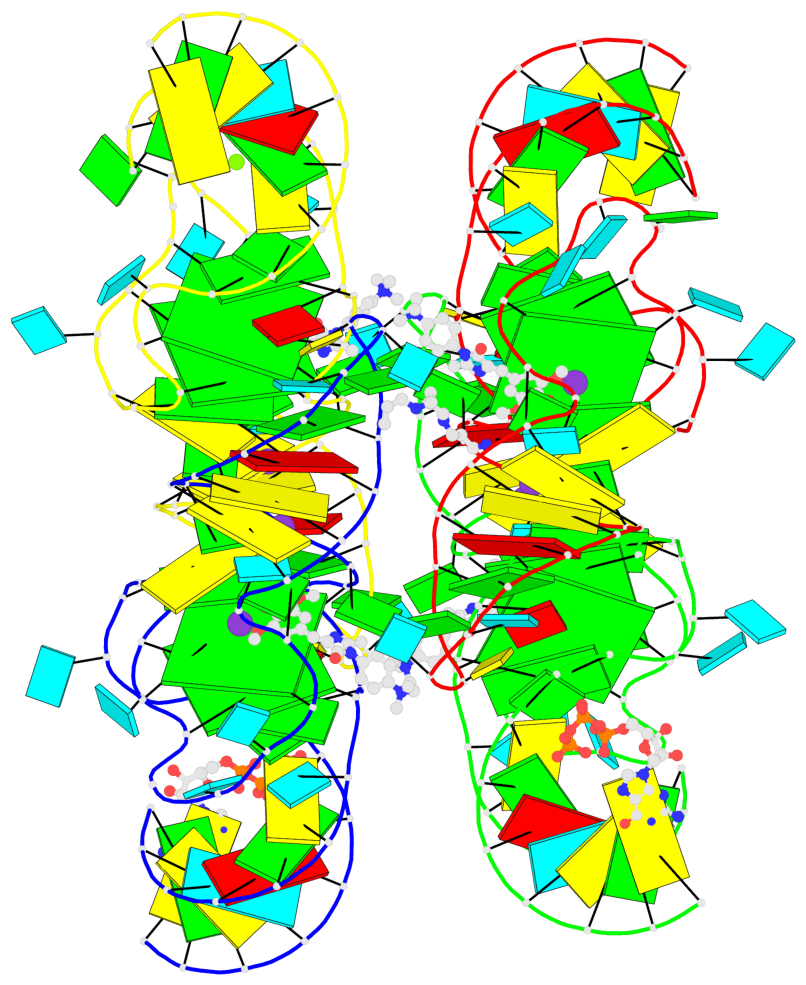
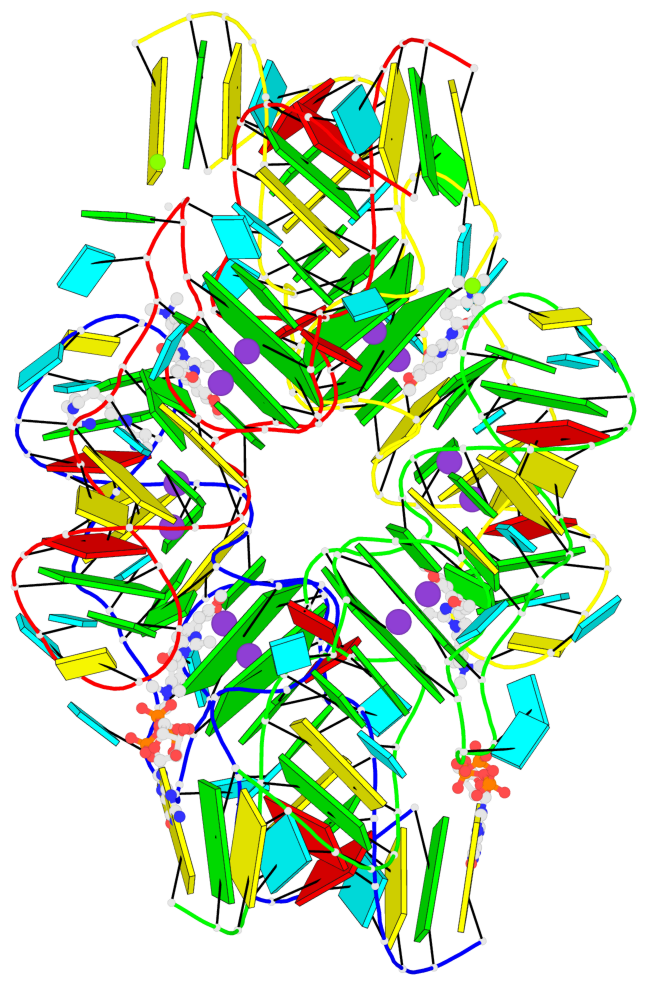
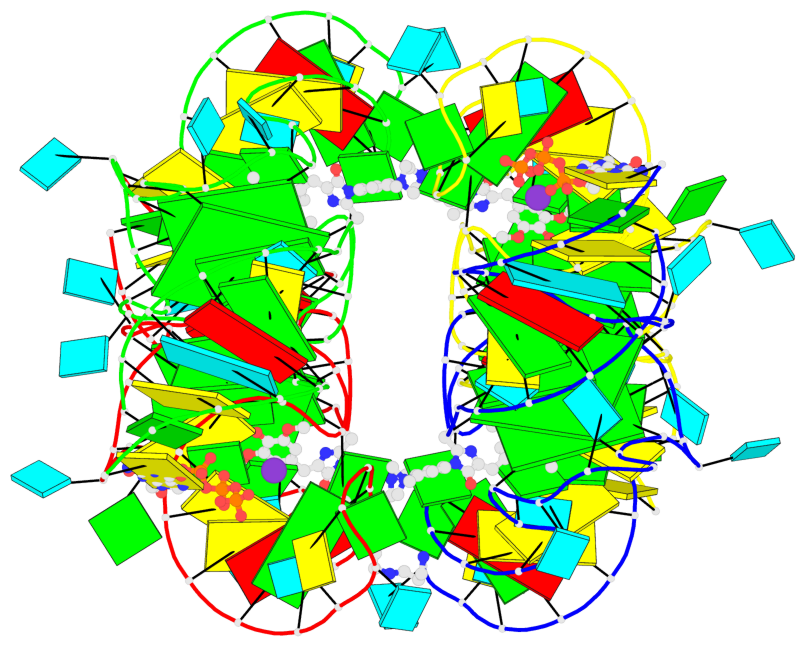
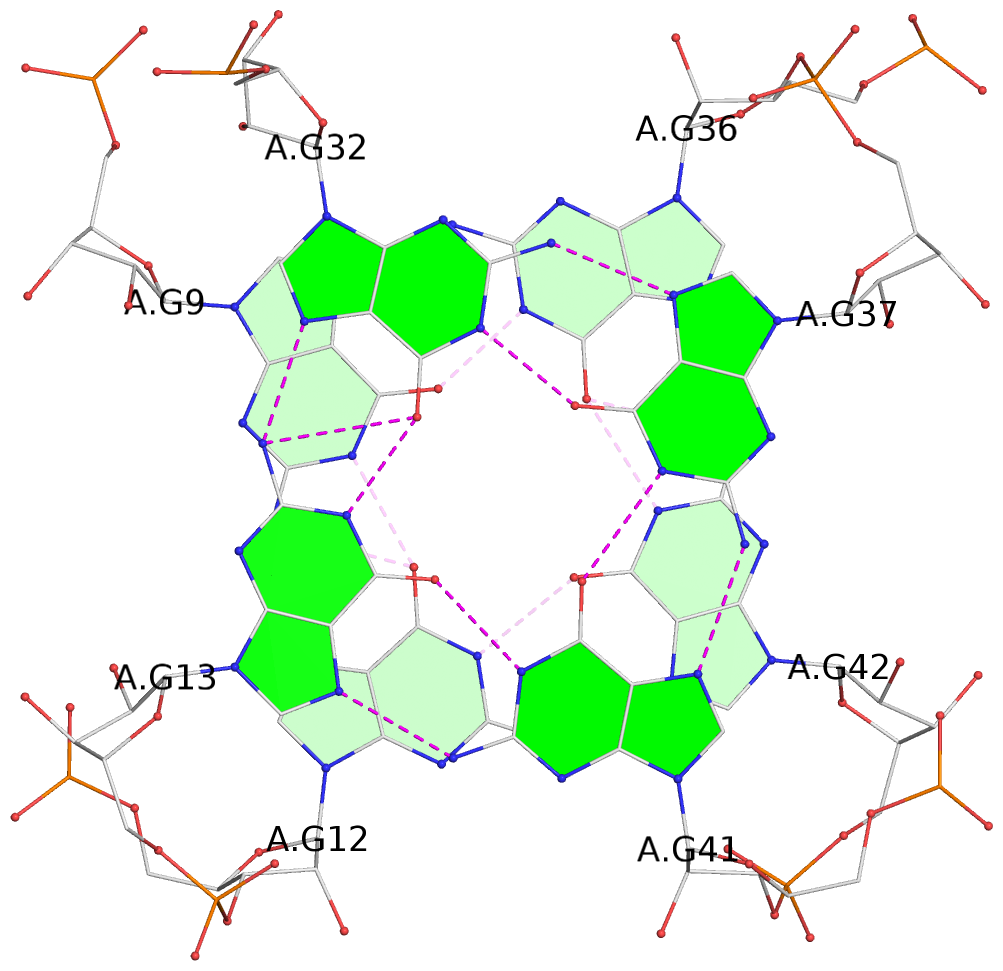
Detailed DSSR results for the G-quadruplex: PDB entry 7oaw
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7oaw
- Class
- RNA
- Method
- X-ray (2.95 Å)
- Summary
- Crystal structure of the chili RNA aptamer in complex with dmhbi+
- Reference
- Mieczkowski M, Steinmetzger C, Bessi I, Lenz AK, Schmiedel A, Holzapfel M, Lambert C, Pena V, Hobartner C (2021): "Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine." Nat Commun, 12, 3549. doi: 10.1038/s41467-021-23932-0.
- Abstract
- Fluorogenic RNA aptamers are synthetic functional RNAs that specifically bind and activate conditional fluorophores. The Chili RNA aptamer mimics large Stokes shift fluorescent proteins and exhibits high affinity for 3,5-dimethoxy-4-hydroxybenzylidene imidazolone (DMHBI) derivatives to elicit green or red fluorescence emission. Here, we elucidate the structural and mechanistic basis of fluorescence activation by crystallography and time-resolved optical spectroscopy. Two co-crystal structures of the Chili RNA with positively charged DMHBO+ and DMHBI+ ligands revealed a G-quadruplex and a trans-sugar-sugar edge G:G base pair that immobilize the ligand by π-π stacking. A Watson-Crick G:C base pair in the fluorophore binding site establishes a short hydrogen bond between the N7 of guanine and the phenolic OH of the ligand. Ultrafast excited state proton transfer (ESPT) from the neutral chromophore to the RNA was found with a time constant of 130 fs and revealed the mode of action of the large Stokes shift fluorogenic RNA aptamer.
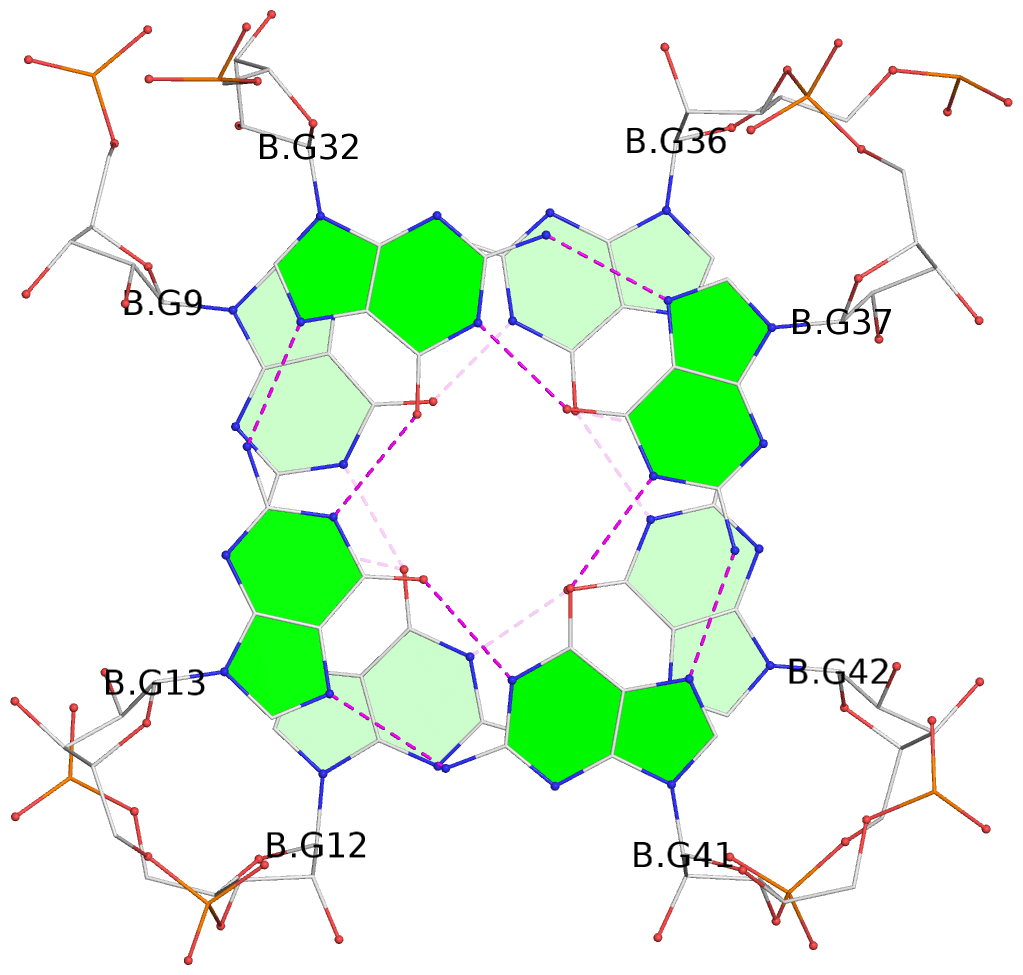
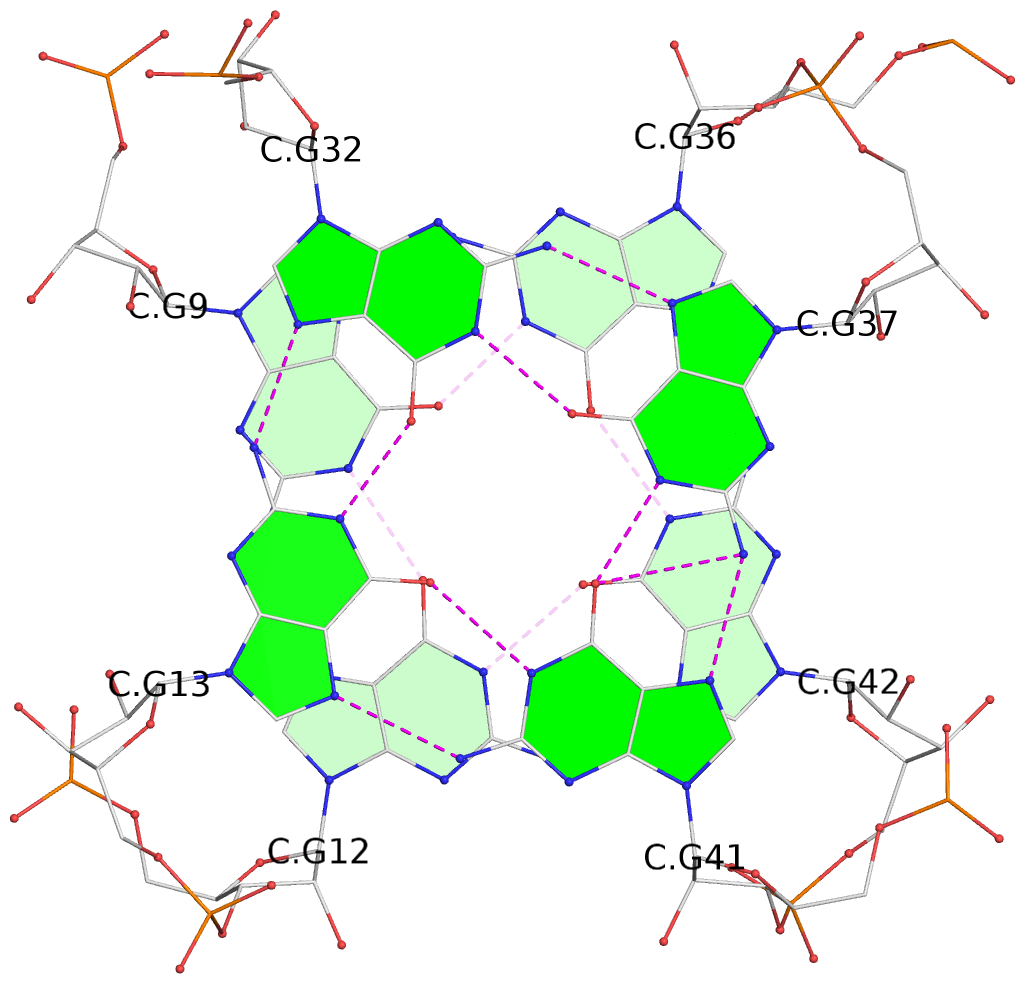
- G4 notes
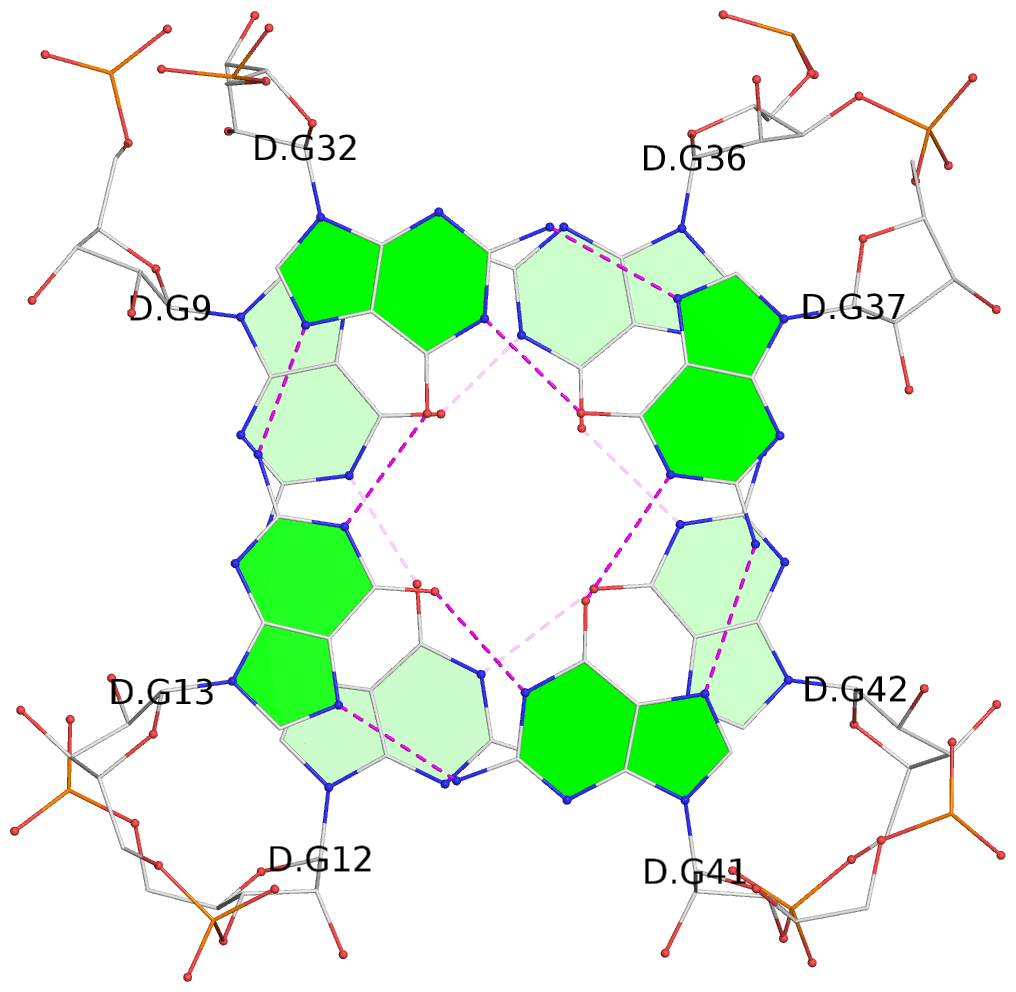
- 8 G-tetrads, 4 G4 helices
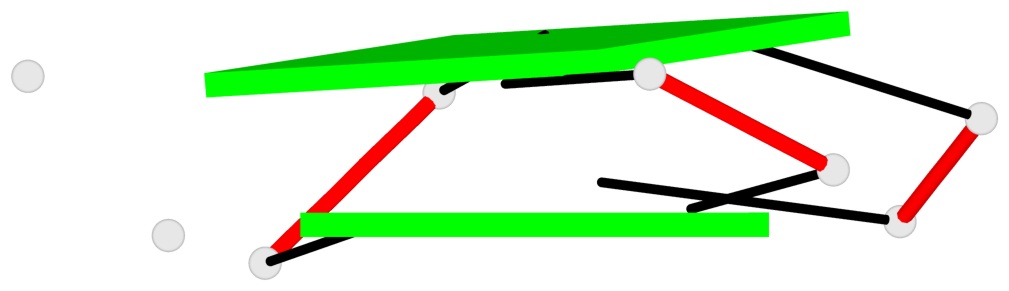
Base-block schematics in six views
List of 8 G-tetrads
1 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.251 type=other nts=4 GGGG A.G9,A.G12,A.G42,A.G36 2 glyco-bond=---- sugar=---3 groove=---- planarity=0.172 type=other nts=4 GGGG A.G13,A.G32,A.G37,A.G41 3 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.281 type=other nts=4 GGGG B.G9,B.G12,B.G42,B.G36 4 glyco-bond=---- sugar=---3 groove=---- planarity=0.183 type=other nts=4 GGGG B.G13,B.G32,B.G37,B.G41 5 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.287 type=other nts=4 GGGG C.G9,C.G12,C.G42,C.G36 6 glyco-bond=---- sugar=---3 groove=---- planarity=0.191 type=other nts=4 GGGG C.G13,C.G32,C.G37,C.G41 7 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.318 type=other nts=4 GGGG D.G9,D.G12,D.G42,D.G36 8 glyco-bond=---- sugar=---3 groove=---- planarity=0.233 type=other nts=4 GGGG D.G13,D.G32,D.G37,D.G41
List of 4 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular
Helix#2, 2 G-tetrad layers, INTRA-molecular
Helix#3, 2 G-tetrad layers, INTRA-molecular
Helix#4, 2 G-tetrad layers, INTRA-molecular
List of 16 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=4 GGAG A.G9,A.G10,A.A11,A.G12 2 type=lateral helix=#1 nts=20 GGGCGCCAGUUCGCUGGUGG A.G13,A.G14,A.G15,A.C16,A.G17,A.C18,A.C19,A.A20,A.G21,A.U22,A.U23,A.C24,A.G25,A.C26,A.U27,A.G28,A.G29,A.U30,A.G31,A.G32 3 type=V-shaped helix=#1 nts=6 GUUGGG A.G32,A.U33,A.U34,A.G35,A.G36,A.G37 4 type=lateral helix=#1 nts=5 GUGCG A.G37,A.U38,A.G39,A.C40,A.G41 5 type=lateral helix=#2 nts=4 GGAG B.G9,B.G10,B.A11,B.G12 6 type=lateral helix=#2 nts=20 GGGCGCCAGUUCGCUGGUGG B.G13,B.G14,B.G15,B.C16,B.G17,B.C18,B.C19,B.A20,B.G21,B.U22,B.U23,B.C24,B.G25,B.C26,B.U27,B.G28,B.G29,B.U30,B.G31,B.G32 7 type=V-shaped helix=#2 nts=6 GUUGGG B.G32,B.U33,B.U34,B.G35,B.G36,B.G37 8 type=lateral helix=#2 nts=5 GUGCG B.G37,B.U38,B.G39,B.C40,B.G41 9 type=lateral helix=#3 nts=4 GGAG C.G9,C.G10,C.A11,C.G12 10 type=lateral helix=#3 nts=20 GGGCGCCAGUUCGCUGGUGG C.G13,C.G14,C.G15,C.C16,C.G17,C.C18,C.C19,C.A20,C.G21,C.U22,C.U23,C.C24,C.G25,C.C26,C.U27,C.G28,C.G29,C.U30,C.G31,C.G32 11 type=V-shaped helix=#3 nts=6 GUUGGG C.G32,C.U33,C.U34,C.G35,C.G36,C.G37 12 type=lateral helix=#3 nts=5 GUGCG C.G37,C.U38,C.G39,C.C40,C.G41 13 type=lateral helix=#4 nts=4 GGAG D.G9,D.G10,D.A11,D.G12 14 type=lateral helix=#4 nts=20 GGGCGCCAGUUCGCUGGUGG D.G13,D.G14,D.G15,D.C16,D.G17,D.C18,D.C19,D.A20,D.G21,D.U22,D.U23,D.C24,D.G25,D.C26,D.U27,D.G28,D.G29,D.U30,D.G31,D.G32 15 type=V-shaped helix=#4 nts=6 GUUGGG D.G32,D.U33,D.U34,D.G35,D.G36,D.G37 16 type=lateral helix=#4 nts=5 GUGCG D.G37,D.U38,D.G39,D.C40,D.G41