Detailed DSSR results for the G-quadruplex: PDB entry 7oqt
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7oqt
- Class
- DNA
- Method
- NMR
- Summary
- G-quadruplex structure of the c. elegans telomeric repeat: a two tetrads basket type conformation stabilised by a hoogsteen c-t base-pair
- Reference
- Marquevielle J, De Rache A, Vialet B, Morvan E, Mergny JL, Amrane S (2022): "G-quadruplex structure of the C. elegans telomeric repeat: a two tetrads basket type conformation stabilized by a non-canonical C-T base-pair." Nucleic Acids Res., 50, 7134-7146. doi: 10.1093/nar/gkac523.
- Abstract
- The Caenorhabditis elegans model has greatly contributed to the understanding of the role of G-quadruplexes in genomic instability. The GGCTTA repeats of the C. elegans telomeres resemble the GGGTTA repeats of the human telomeres. However, the comparison of telomeric sequences (Homo sapiens, Tetrahymena, Oxytricha, Bombyx mori and Giardia) revealed that small changes in these repeats can drastically change the topology of the folded G-quadruplex. In the present work we determined the structure adopted by the C. elegans telomeric sequence d[GG(CTTAGG)3]. The investigated C. elegans telomeric sequence is shown to fold into an intramolecular two G-tetrads basket type G-quadruplex structure that includes a C-T base pair in the diagonal loop. This work sheds light on the telomeric structure of the widely used C. elegans animal model.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-LwD+Ln), basket(2+2), UDDU
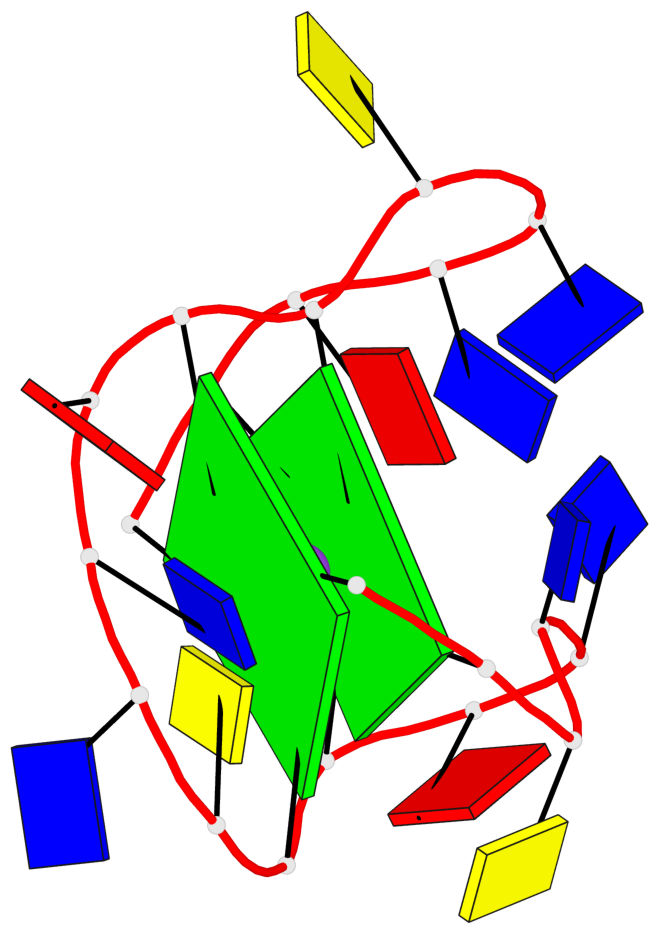
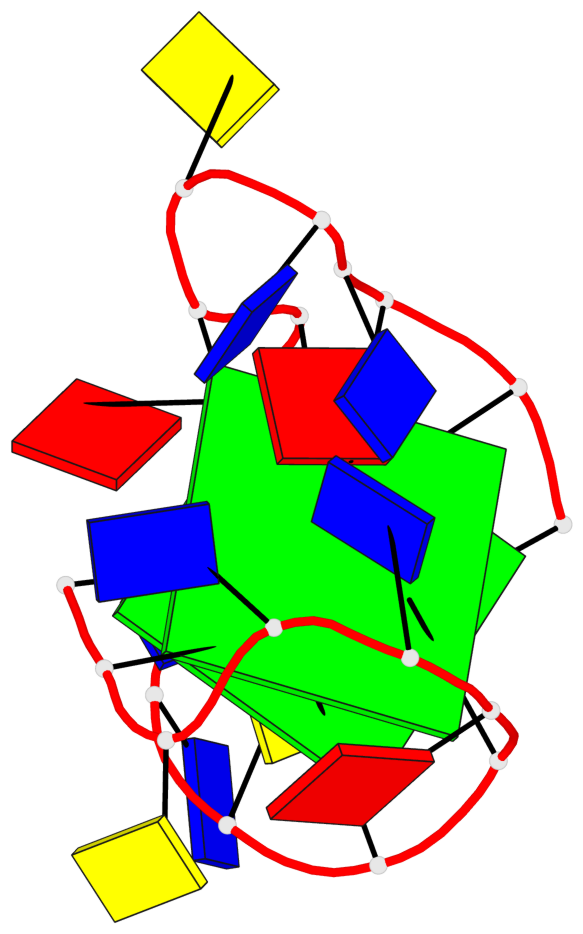
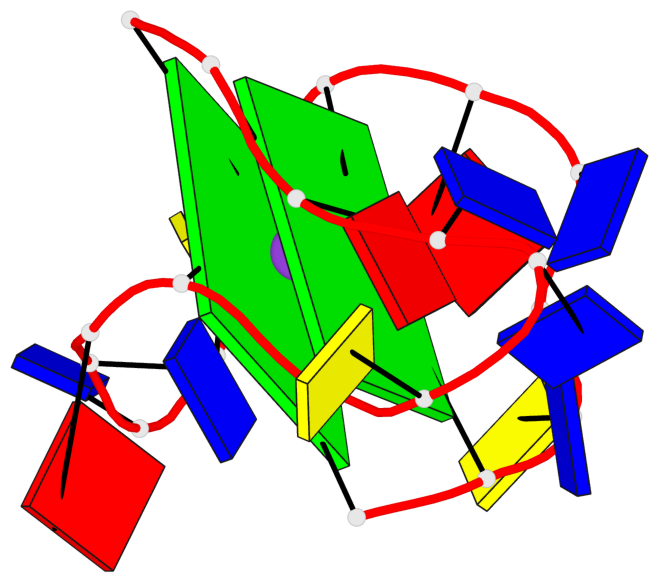
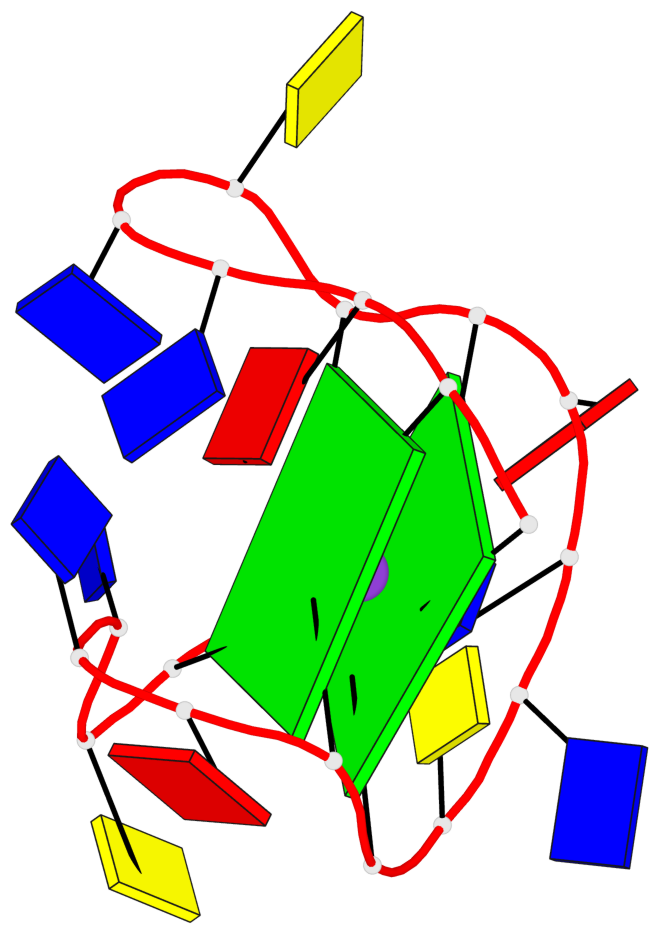
Base-block schematics in six views
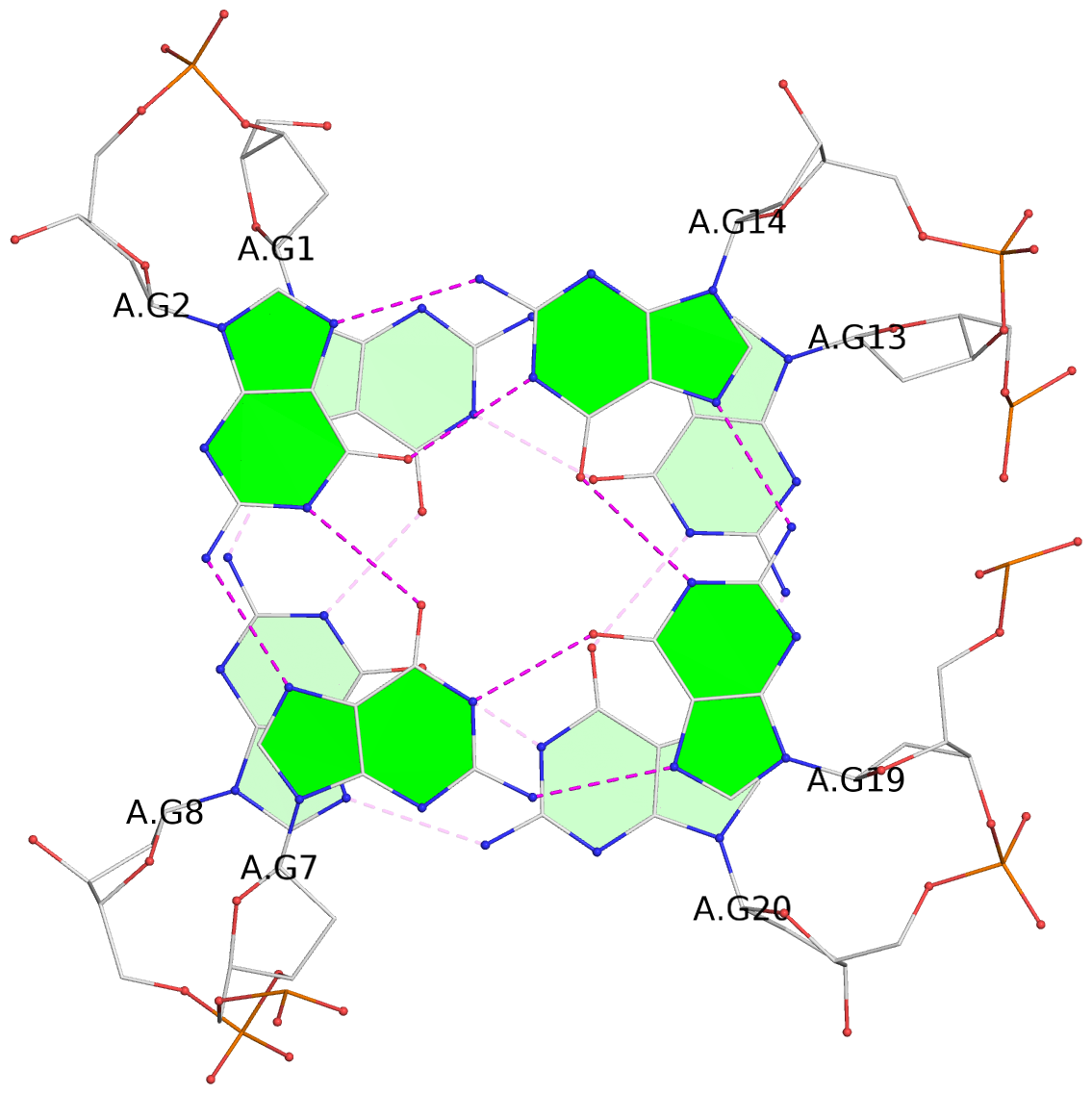
List of 2 G-tetrads
1 glyco-bond=s--s sugar=--.. groove=w-n- planarity=0.224 type=other nts=4 GGGG A.DG1,A.DG8,A.DG20,A.DG13 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.184 type=other nts=4 GGGG A.DG2,A.DG7,A.DG19,A.DG14
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.