Detailed DSSR results for the G-quadruplex: PDB entry 7otb
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7otb
- Class
- DNA
- Method
- X-ray (1.6 Å)
- Summary
- Ruthenium polypridyl complex bound to a unimolecular chair-form g-quadruplex
- Reference
- McQuaid KT, Takahashi S, Baumgaertner L, Cardin DJ, Paterson NG, Hall JP, Sugimoto N, Cardin CJ (2022): "Ruthenium Polypyridyl Complex Bound to a Unimolecular Chair-Form G-Quadruplex." J.Am.Chem.Soc., 144, 5956-5964. doi: 10.1021/jacs.2c00178.
- Abstract
- The DNA G-quadruplex is known for forming a range of topologies and for the observed lability of the assembly, consistent with its transient formation in live cells. The stabilization of a particular topology by a small molecule is of great importance for therapeutic applications. Here, we show that the ruthenium complex Λ-[Ru(phen)2(qdppz)]2+ displays enantiospecific G-quadruplex binding. It crystallized in 1:1 stoichiometry with a modified human telomeric G-quadruplex sequence, GGGTTAGGGTTAGGGTTTGGG (htel21T18), in an antiparallel chair topology, the first structurally characterized example of ligand binding to this topology. The lambda complex is bound in an intercalation cavity created by a terminal G-quartet and the central narrow lateral loop formed by T10-T11-A12. The two remaining wide lateral loops are linked through a third K+ ion at the other end of the G-quartet stack, which also coordinates three thymine residues. In a comparative ligand-binding study, we showed, using a Klenow fragment assay, that this complex is the strongest observed inhibitor of replication, both using the native human telomeric sequence and the modified sequence used in this work.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-Lw), chair(2+2), UDUD
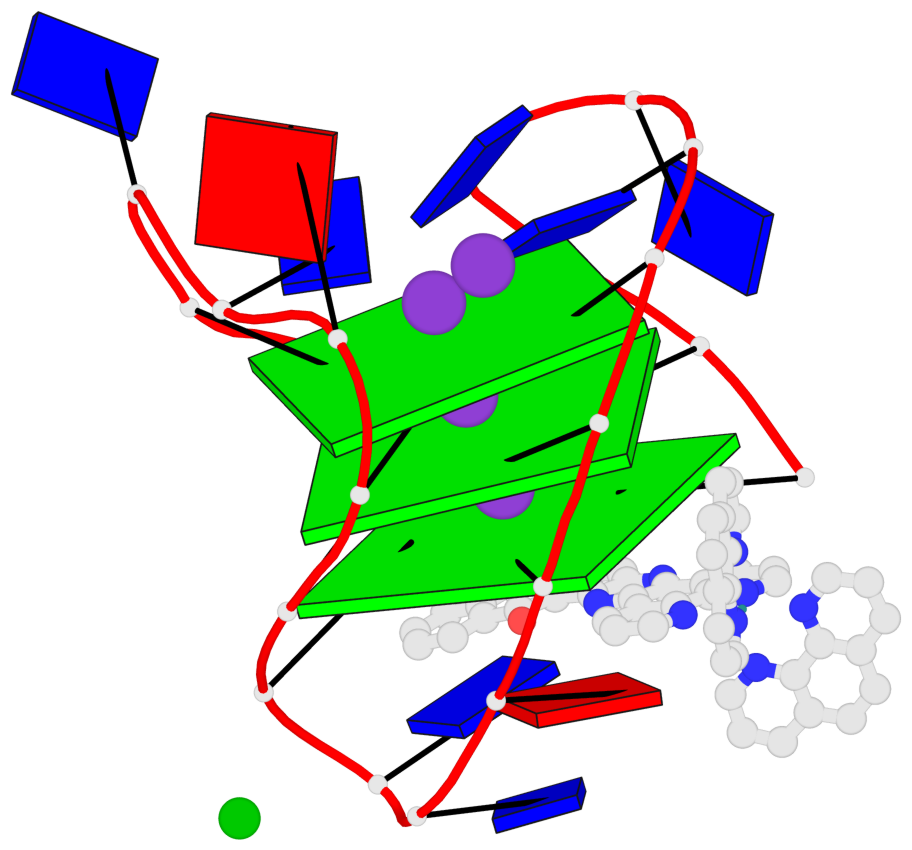
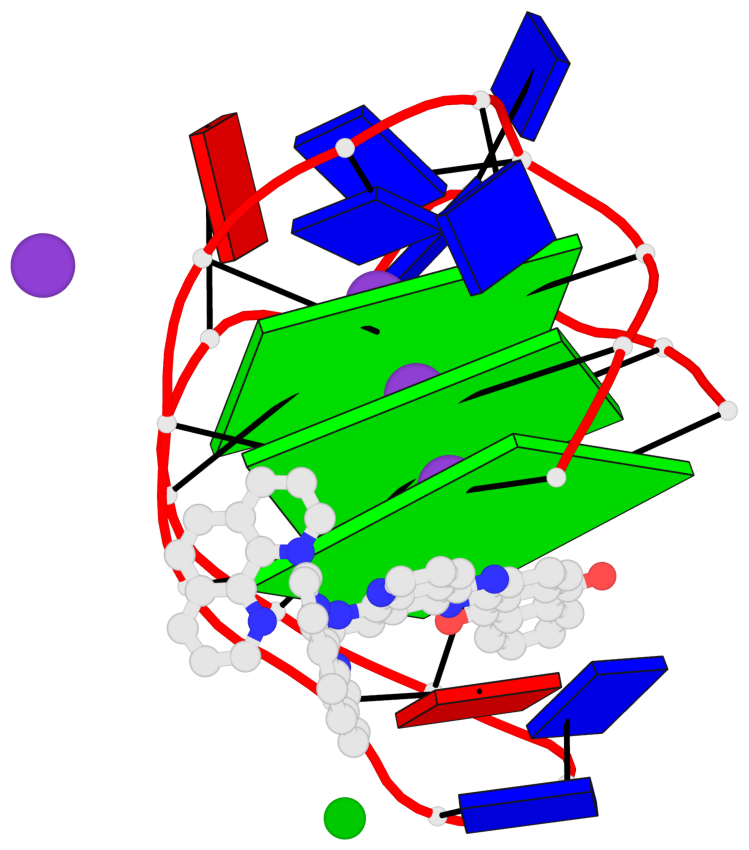
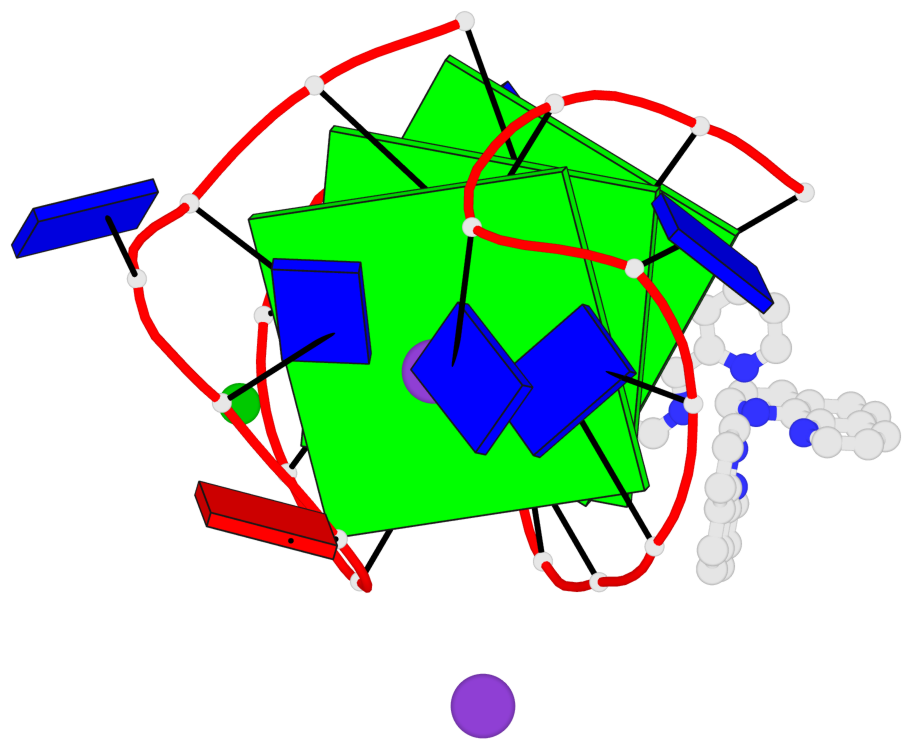
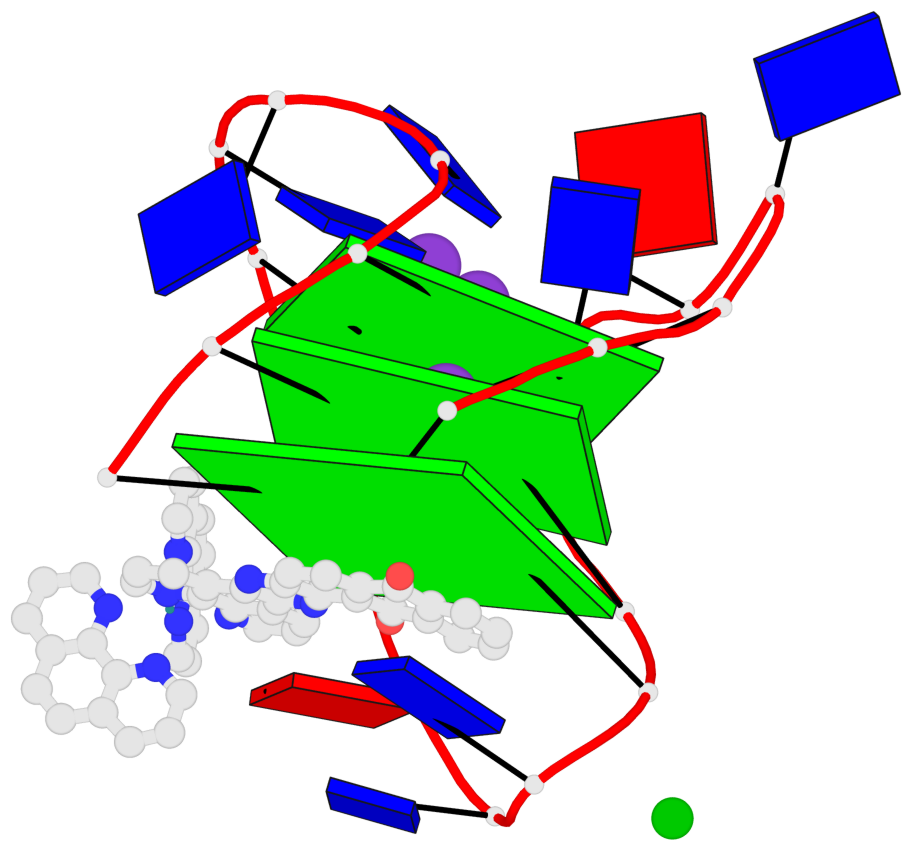
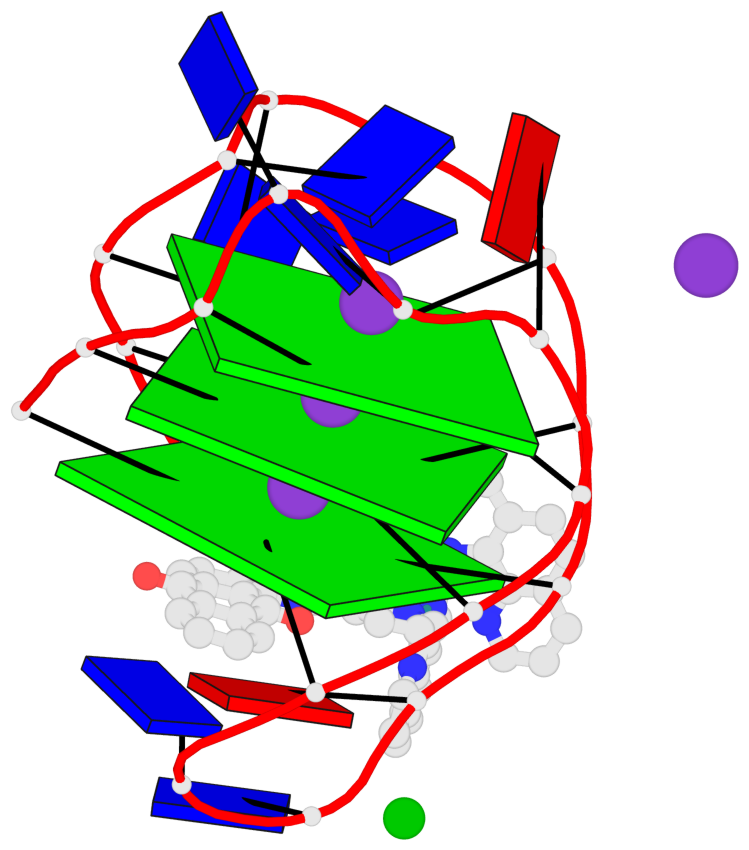
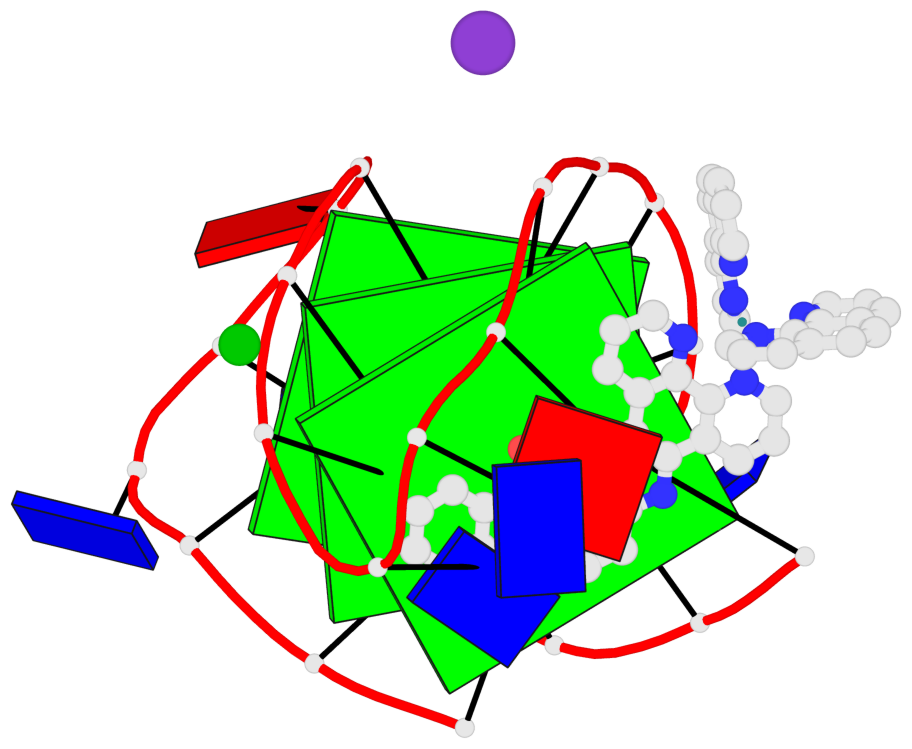
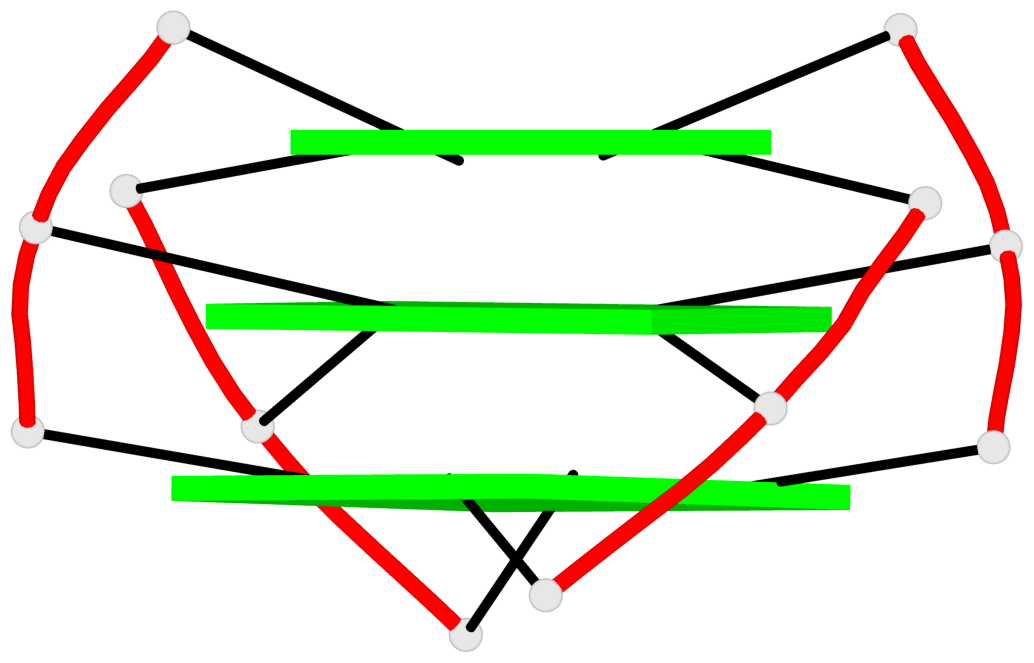
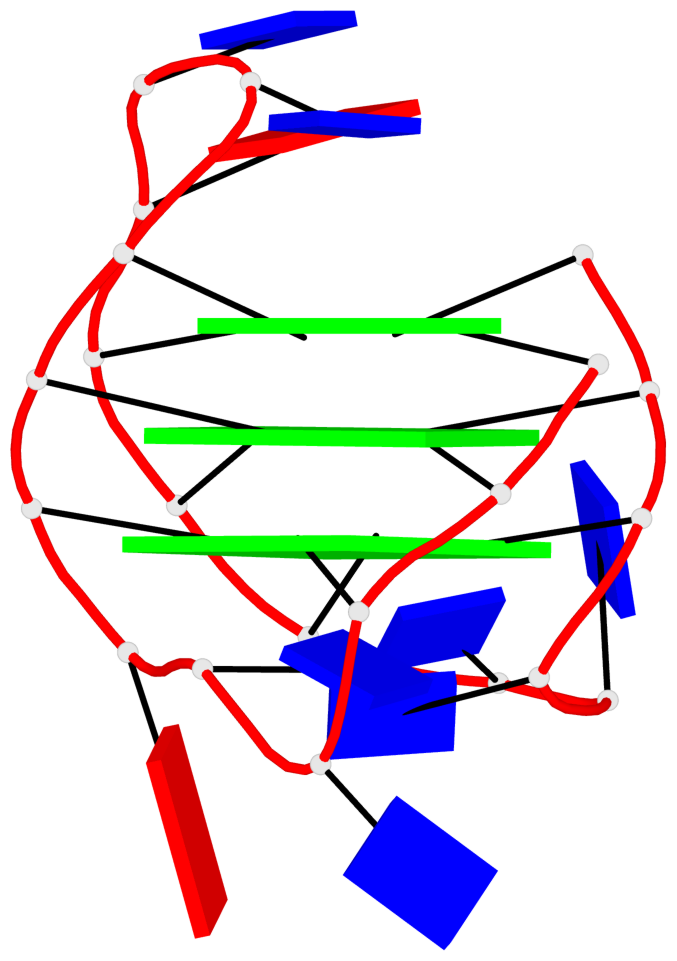
Base-block schematics in six views
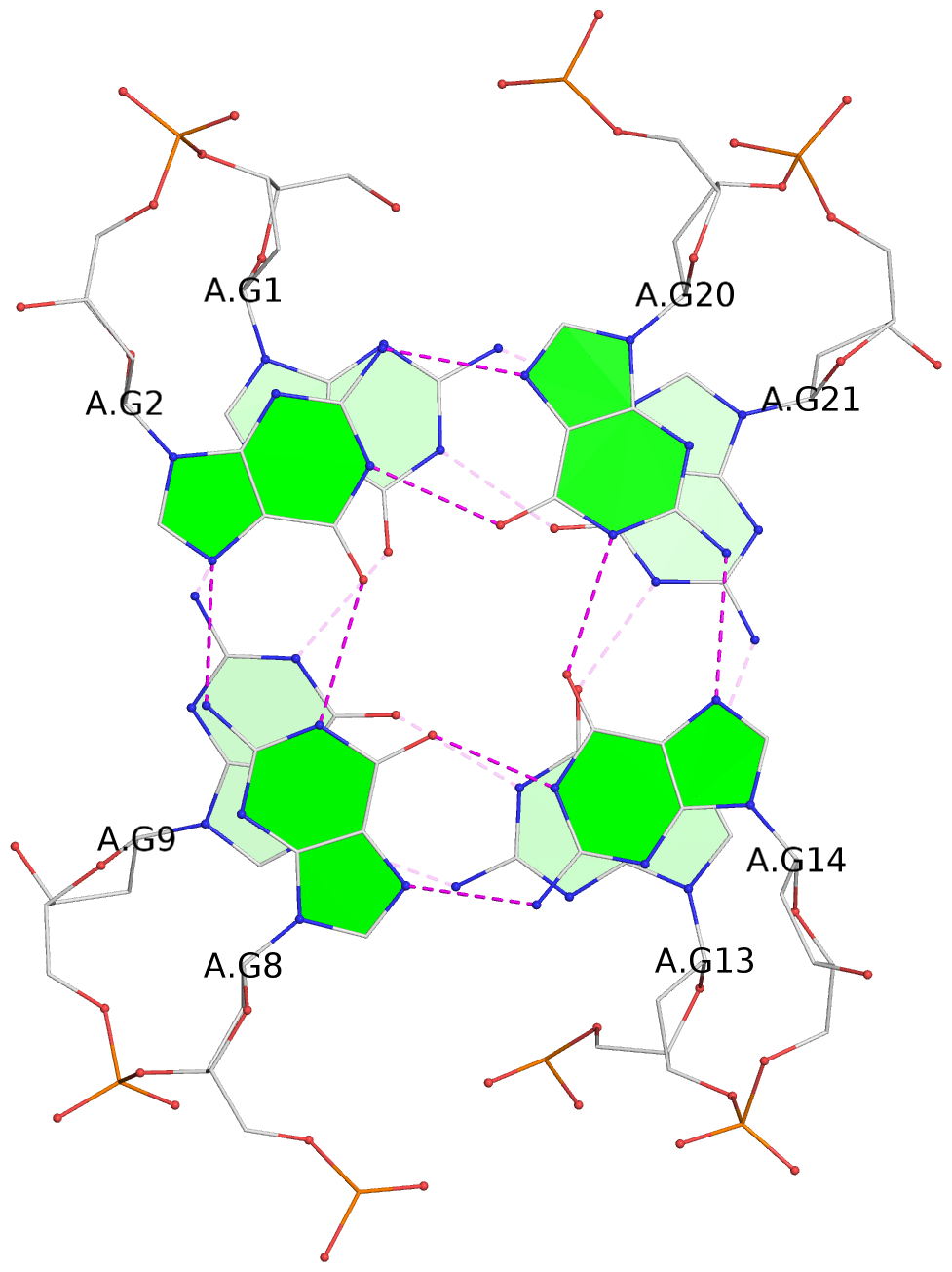
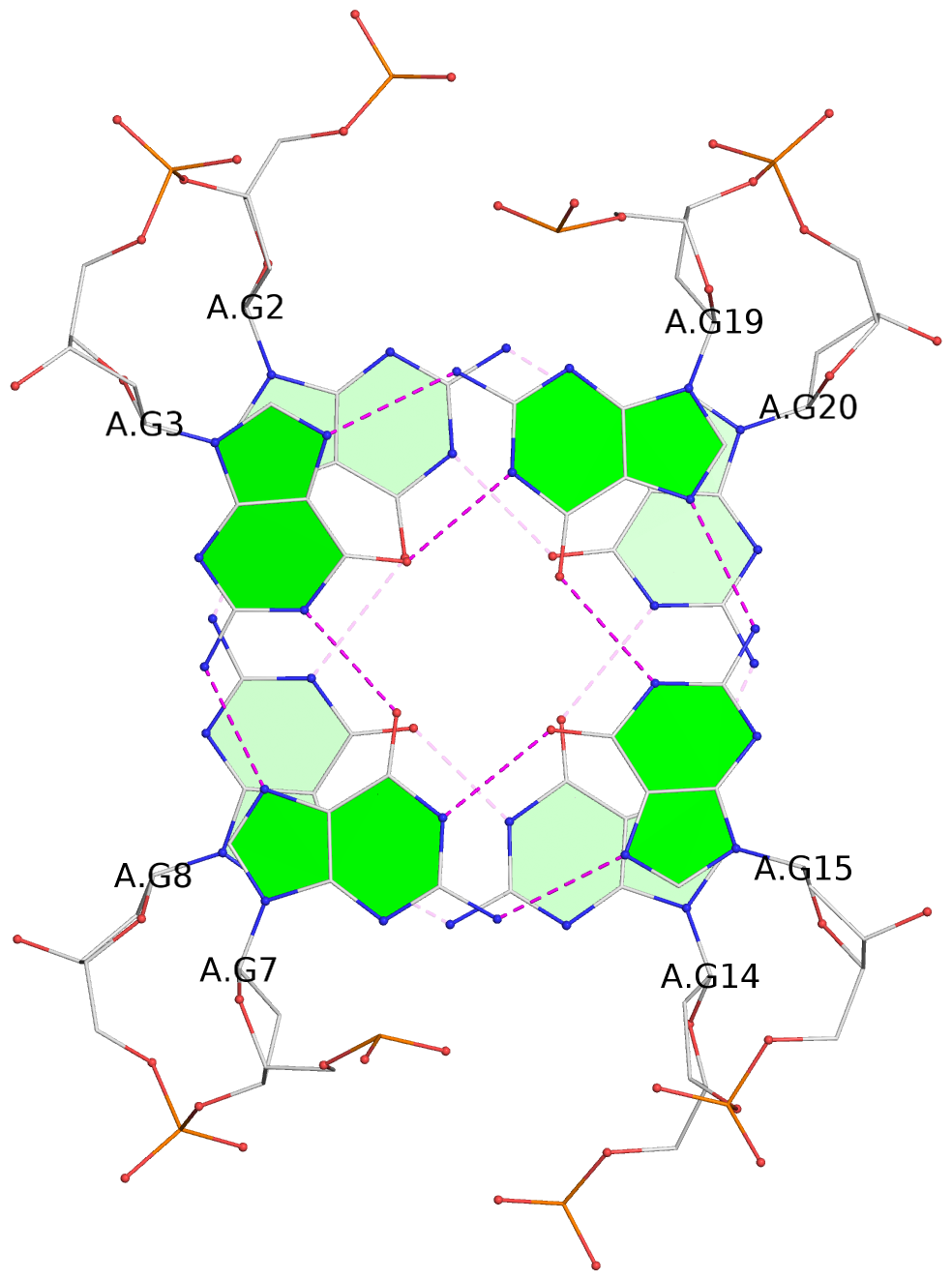
List of 3 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.543 type=bowl-2 nts=4 GGGG A.DG1,A.DG9,A.DG13,A.DG21 2 glyco-bond=s-s- sugar=--.- groove=wnwn planarity=0.515 type=other nts=4 GGGG A.DG2,A.DG8,A.DG14,A.DG20 3 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.568 type=saddle nts=4 GGGG A.DG3,A.DG7,A.DG15,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.