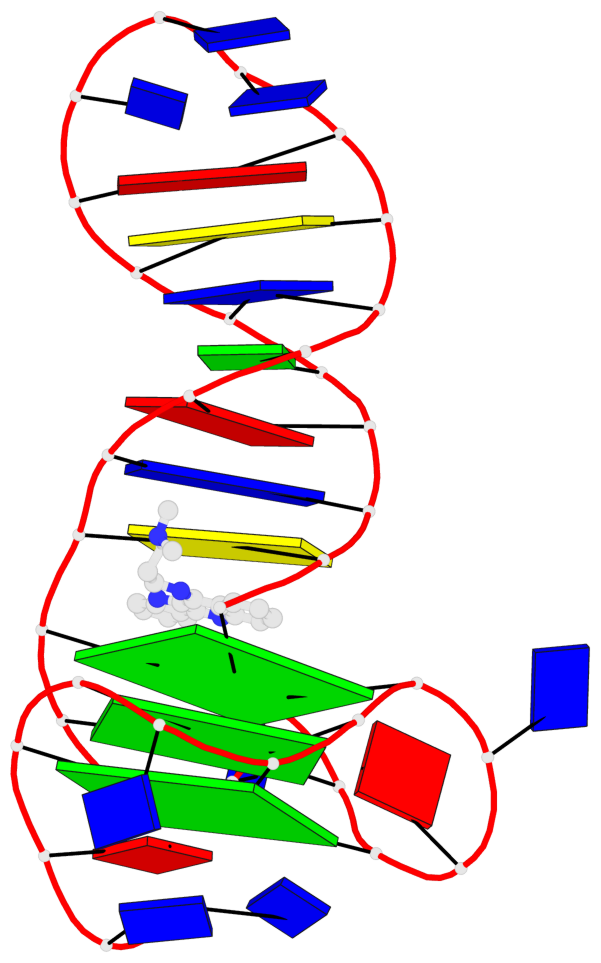
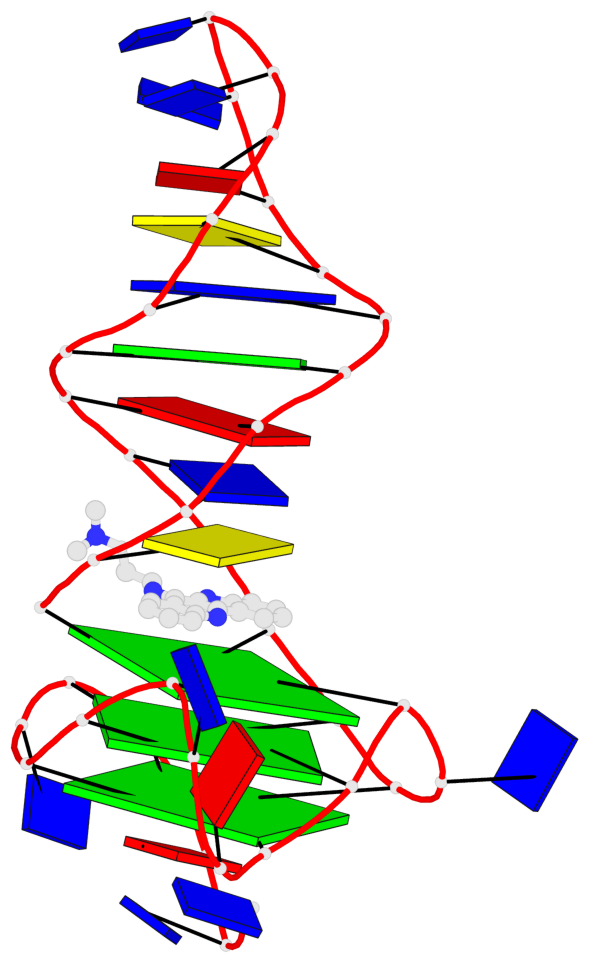
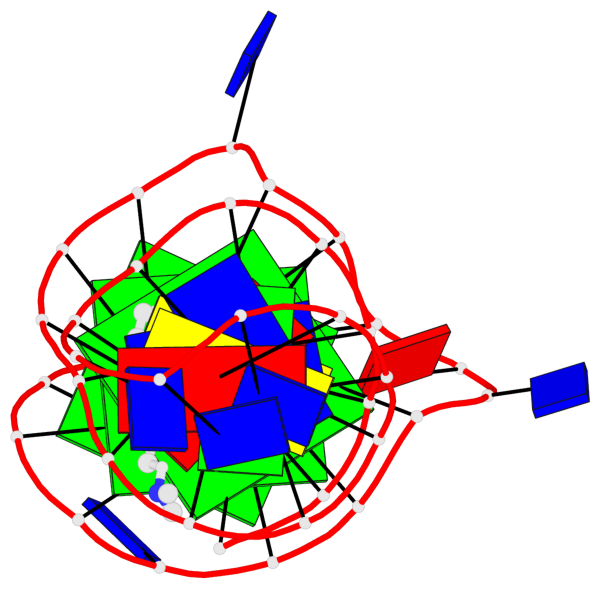
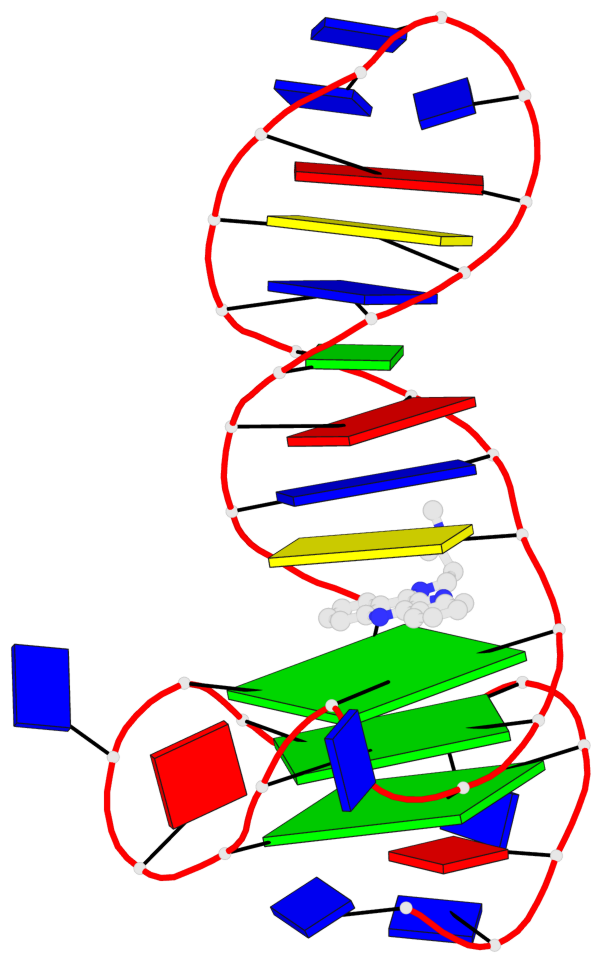
Detailed DSSR results for the G-quadruplex: PDB entry 7png
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7png
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of 1:1 complex of an indoloquinoline derivative syuiq-5 to parallel quadruplex-duplex (q-d) hybrid
- Reference
- Vianney YM, Weisz K (2021): "Indoloquinoline Ligands Favor Intercalation at Quadruplex-Duplex Interfaces." Chemistry, e202103718. doi: 10.1002/chem.202103718.
- Abstract
- Quadruplex-duplex (Q-D) junctions are increasingly considered promising targets for medicinal and technological applications. Here, a Q-D hybrid with a hairpin-type snapback loop coaxially stacked onto the quadruplex 3'-outer tetrad was designed and employed as a target structure for the indoloquinoline ligand SYUIQ-5. NMR spectral analysis demonstrated high-affinity binding of the ligand at the quadruplex-duplex interface with association constants determined by isothermal titration calorimetry of about 10 7 M -1 and large exothermicities Δ H ° of -14 kcal/mol in a 120 mM K + buffer at 40 °C. Determination of the ligand-bound hybrid structure revealed intercalation of SYUIQ-5 between 3'-outer tetrad and the neighboring CG base pair, maximizing π-π stacking as well as electrostatic interactions with guanine carbonyl groups in close vicinity to the positively charged protonated quinoline nitrogen of the tetracyclic indoloquinoline. Exhibiting considerable flexibility, the SYUIQ-5 sidechain resides in the duplex minor groove. Based on comparative binding studies with the non-substituted N 5-methylated indoloquinoline cryptolepine, the sidechain is suggested to confer additional affinity and to fix the alignment of the intercalated indoloquinoline aromatic core. However, selectivity for the Q-D junction mostly relies on the geometry and charge distribution of the indoloquinoline ring system. The presented results are expected to provide valuable guidelines for the design of ligands specifically targeting Q-D interfaces.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
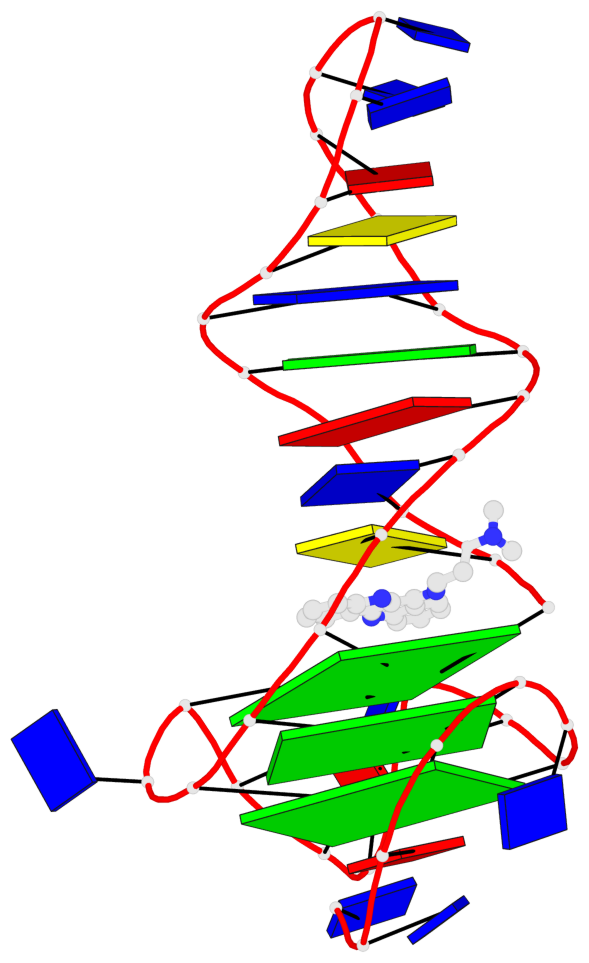
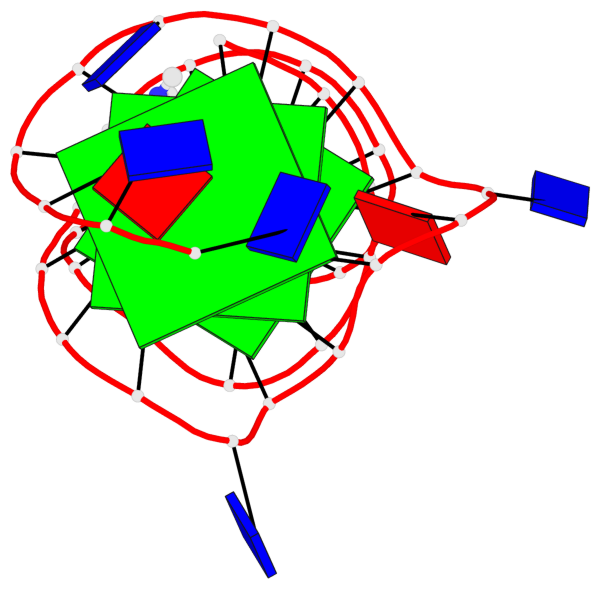
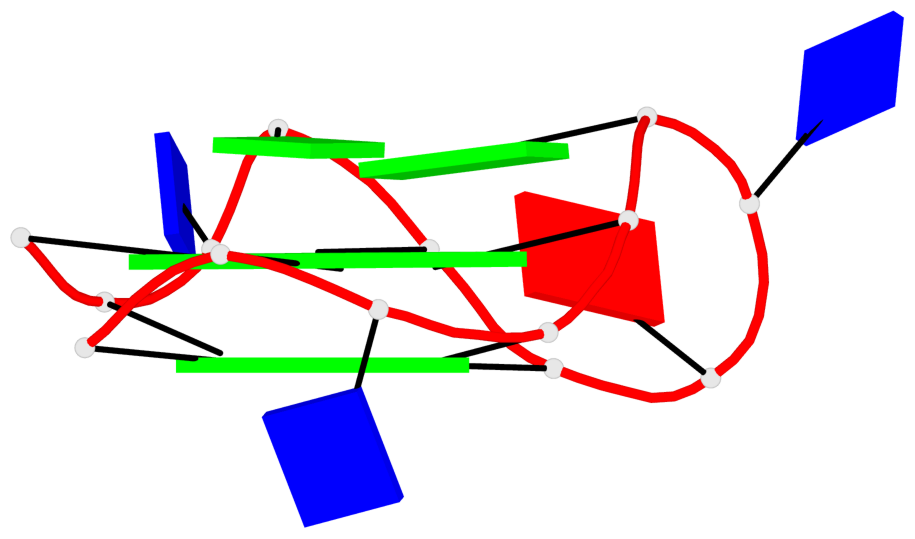
Base-block schematics in six views
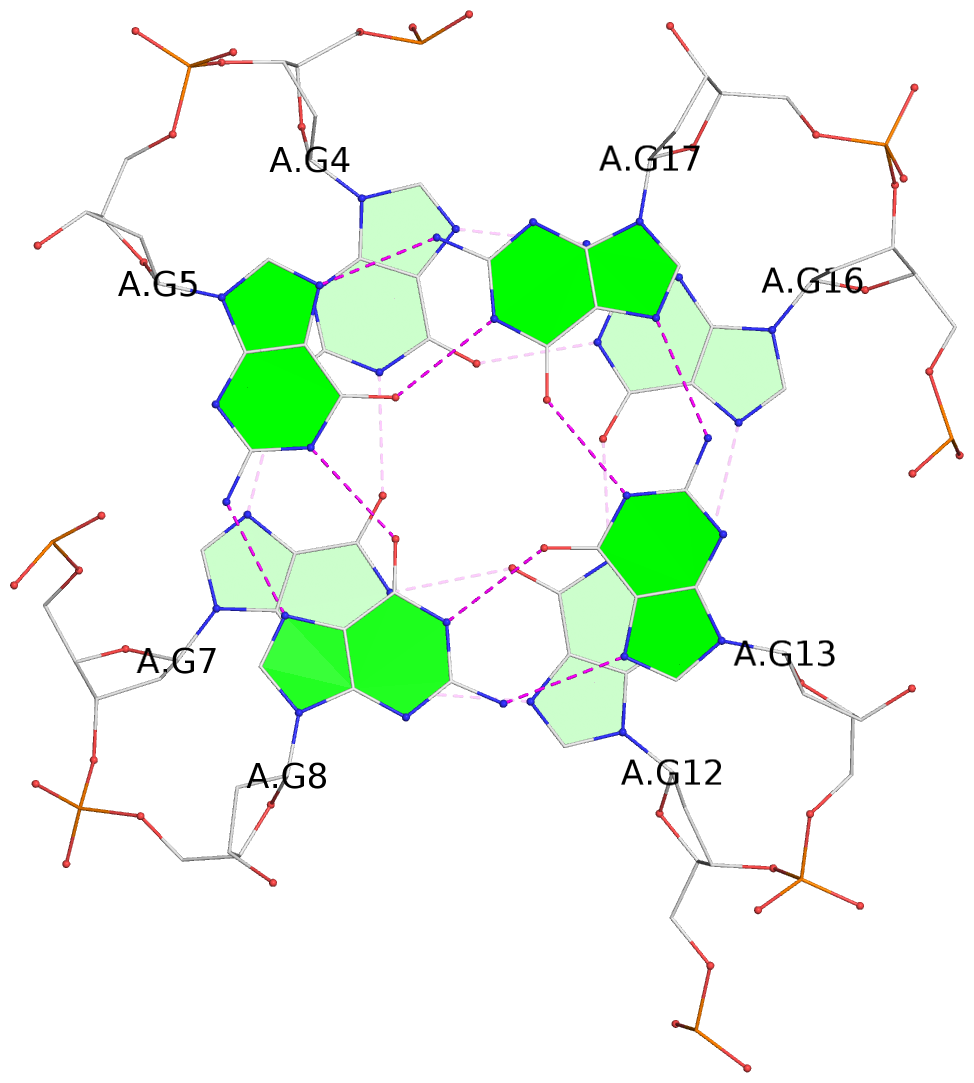
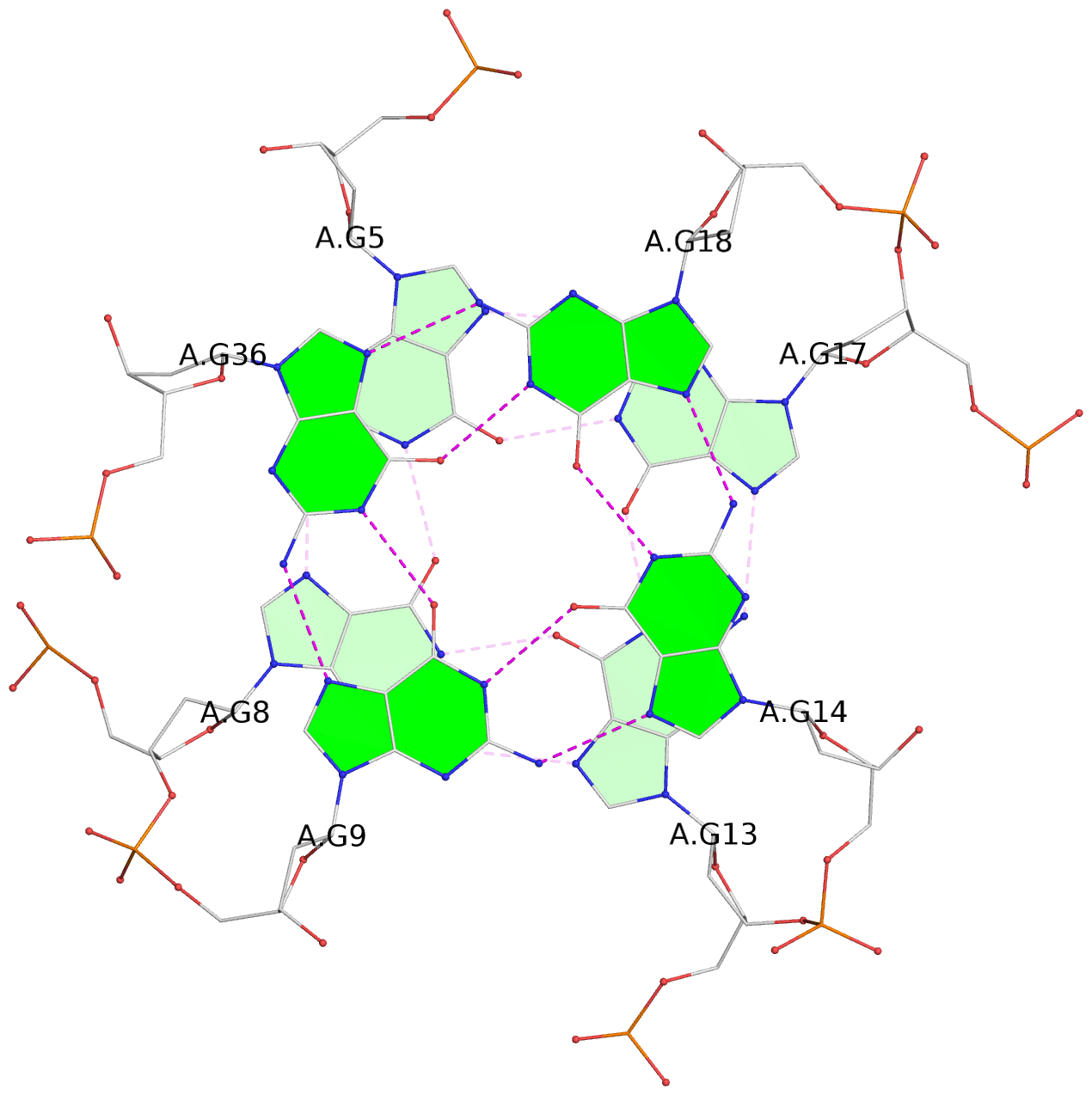
List of 3 G-tetrads
1 glyco-bond=---- sugar=--.- groove=---- planarity=0.261 type=other nts=4 GGGG A.DG4,A.DG7,A.DG12,A.DG16 2 glyco-bond=---- sugar=---- groove=---- planarity=0.235 type=other nts=4 GGGG A.DG5,A.DG8,A.DG13,A.DG17 3 glyco-bond=---s sugar=---- groove=--wn planarity=0.316 type=other nts=4 GGGG A.DG9,A.DG14,A.DG18,A.DG36
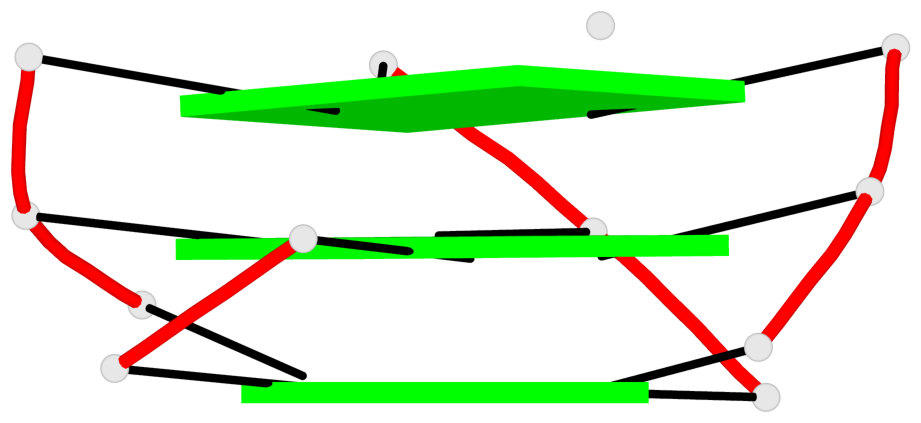
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 non-stem G4-loop (including the two closing Gs)
1 type=lateral helix=#1 nts=19 GCTAGTCATTTTGACTAGG A.DG18,A.DC19,A.DT20,A.DA21,A.DG22,A.DT23,A.DC24,A.DA25,A.DT26,A.DT27,A.DT28,A.DT29,A.DG30,A.DA31,A.DC32,A.DT33,A.DA34,A.DG35,A.DG36