Detailed DSSR results for the G-quadruplex: PDB entry 7x2z
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7x2z
- Class
- DNA
- Method
- NMR
- Summary
- NMR solution structure of the 1:1 complex of a pyridostatin derivative (pypds) bound to a g-quadruplex myt1l
- Reference
- Liu L-Y, Ma TZ, Zeng YL, Liu W, Mao Z-W (2022): "Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes." J.Am.Chem.Soc., 144, 11878-11887. doi: 10.1021/jacs.2c04775.
- Abstract
- The nucleic acid G-quadruplex (G4) has emerged as a promising therapeutic target for a variety of diseases such as cancer and neurodegenerative disease. Among small-molecule G4-binders, pyridostatin (PDS) and its derivatives (e.g., PyPDS) exhibit high specificity to G4s, but the structural basis for their specific recognition of G4s remains unknown. Here, we presented two solution structures of PyPDS and PDS with a quadruplex-duplex hybrid. The structures indicate that the rigid aromatic rings of PyPDS/PDS linked by flexible amide bonds match adaptively with G-tetrad planes, enhancing π-π stacking and achieving specific recognition of G4s. The aliphatic amine side chains of PyPDS/PDS adjust conformation to interact with the phosphate backbone via hydrogen bonding and electrostatic interactions, increasing affinity for G4s. Moreover, the N-H of PyPDS/PDS amide bonds interacts with two O6s of G-tetrad guanines via hydrogen bonding, achieving a further increase in affinity for G4s, which is different from most G4 ligands. Our findings reveal from structural perspectives that the rational assembly of rigid and flexible structural units in a ligand can synergistically improve the selectivity and affinity for G4s through spatial selective and adaptive matching.
- G4 notes
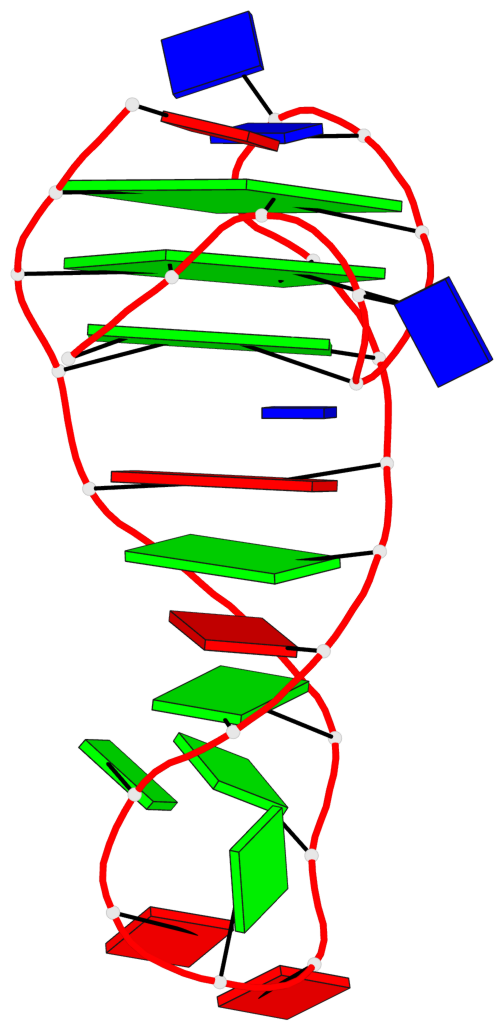
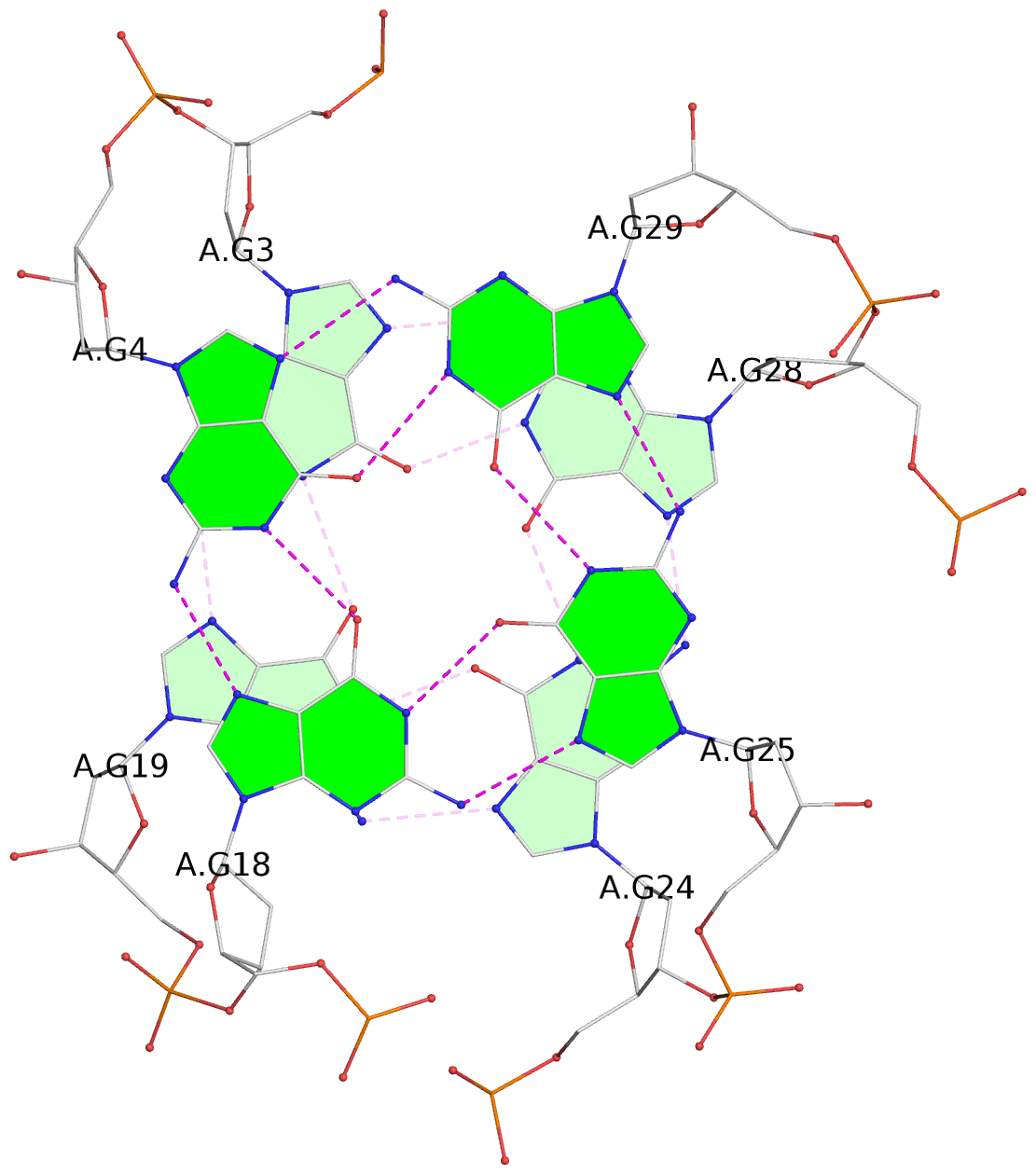
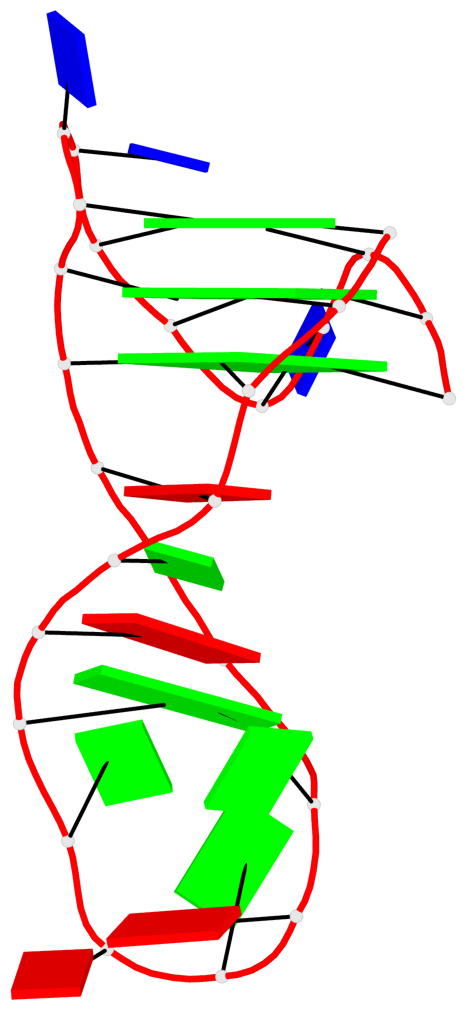
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
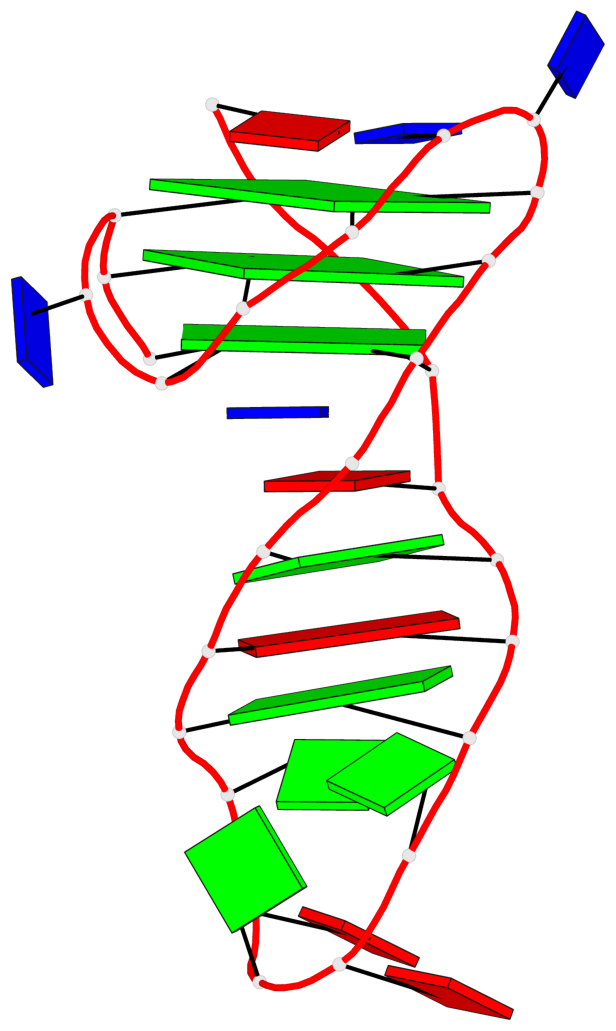
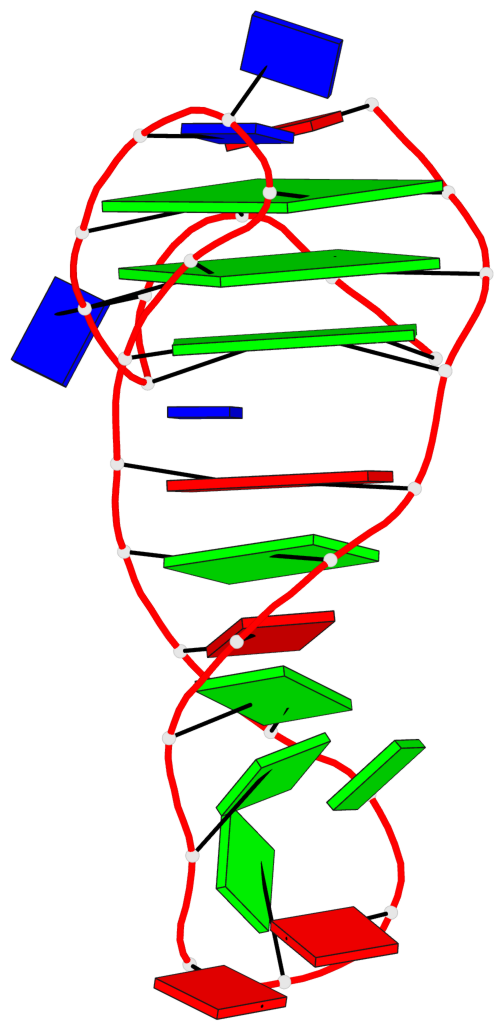
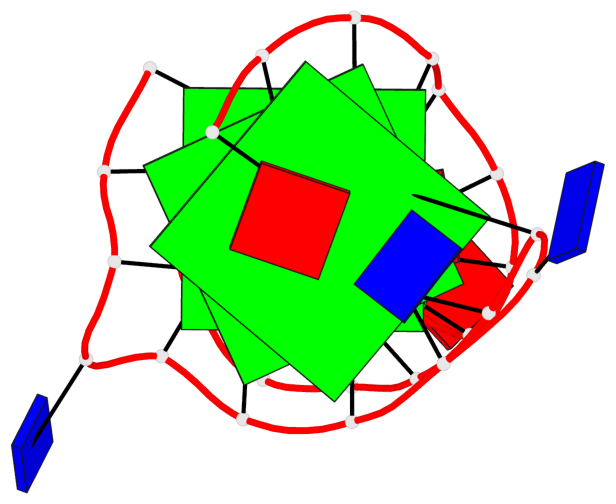
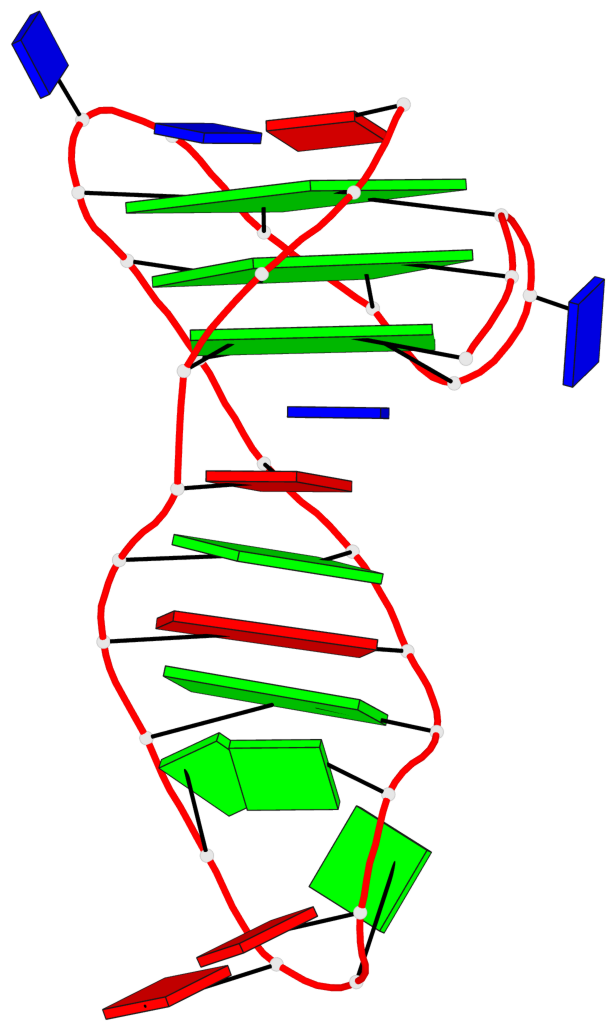
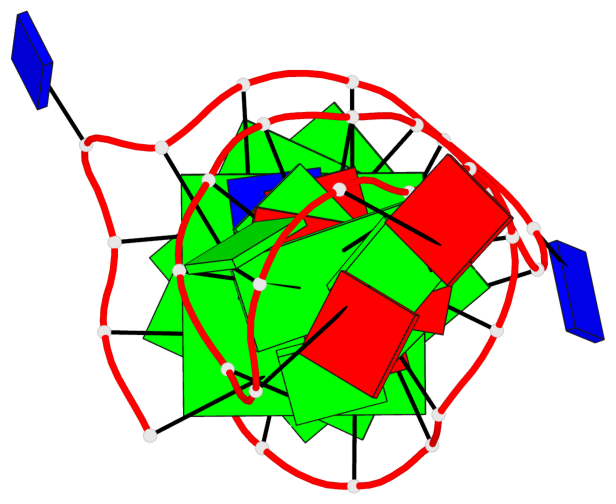
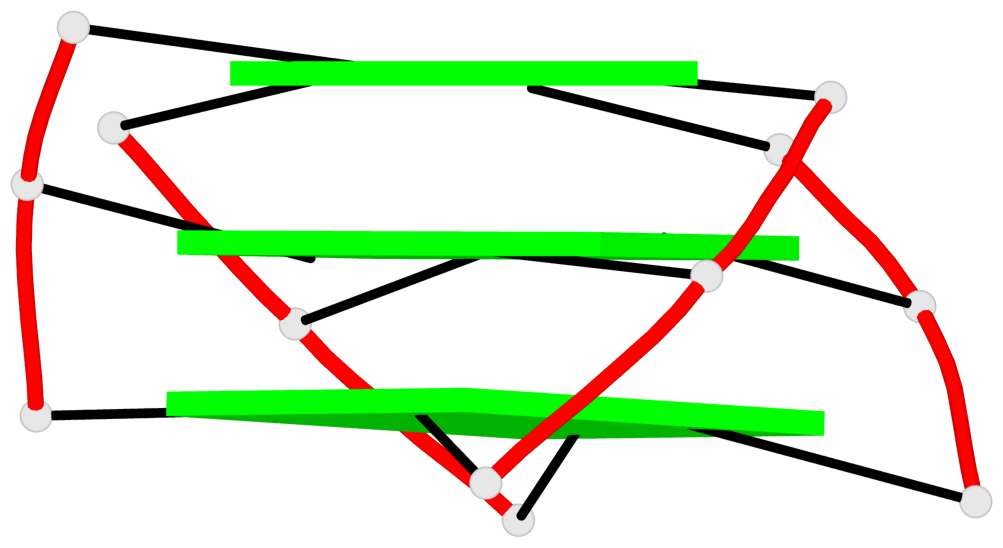
Base-block schematics in six views
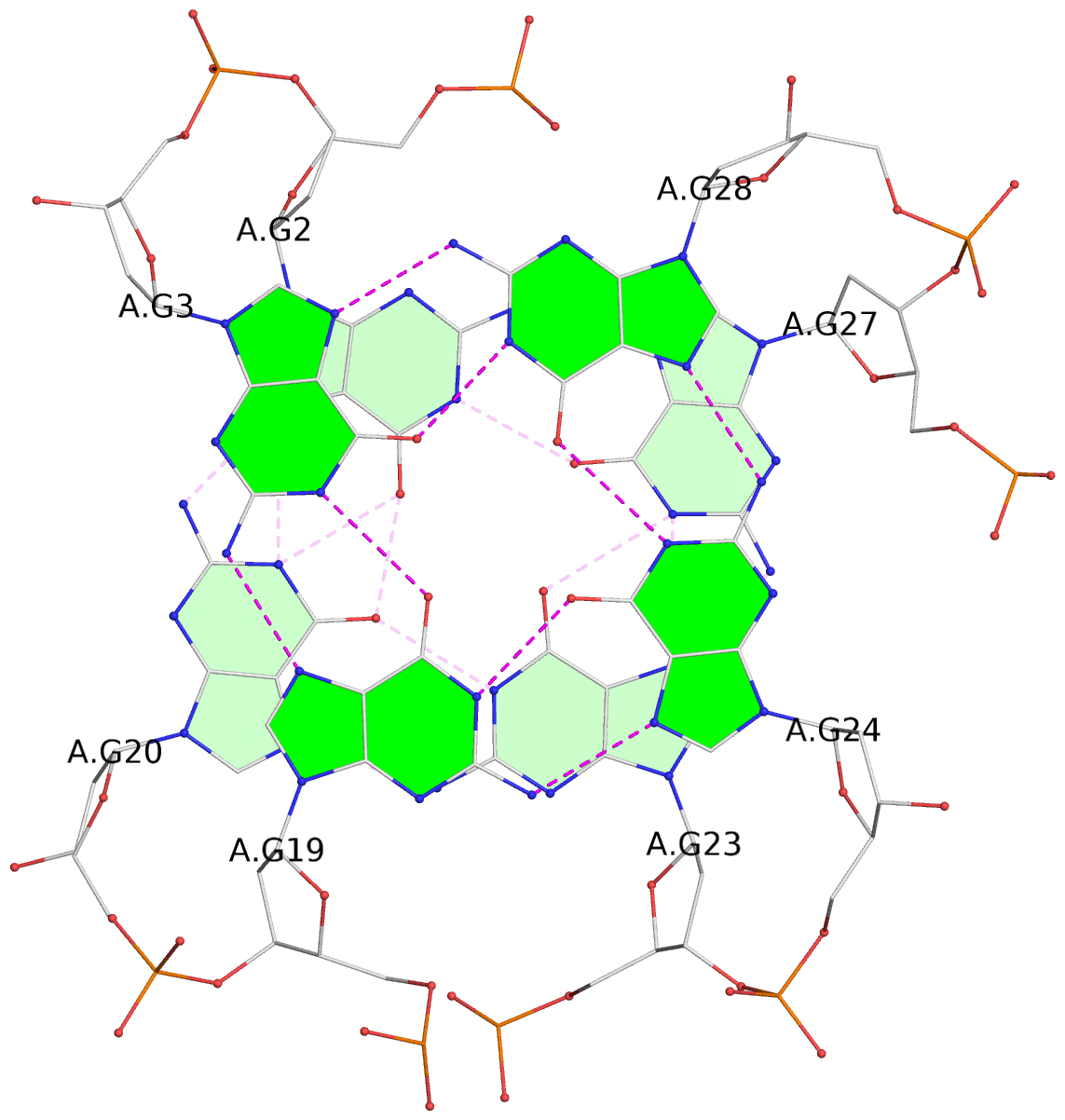
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=.... groove=wn-- planarity=0.161 type=other nts=4 GGGG A.DG2,A.DG20,A.DG23,A.DG27 2 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.110 type=planar nts=4 GGGG A.DG3,A.DG19,A.DG24,A.DG28 3 glyco-bond=-s-- sugar=3-.. groove=wn-- planarity=0.097 type=planar nts=4 GGGG A.DG4,A.DG18,A.DG25,A.DG29
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.