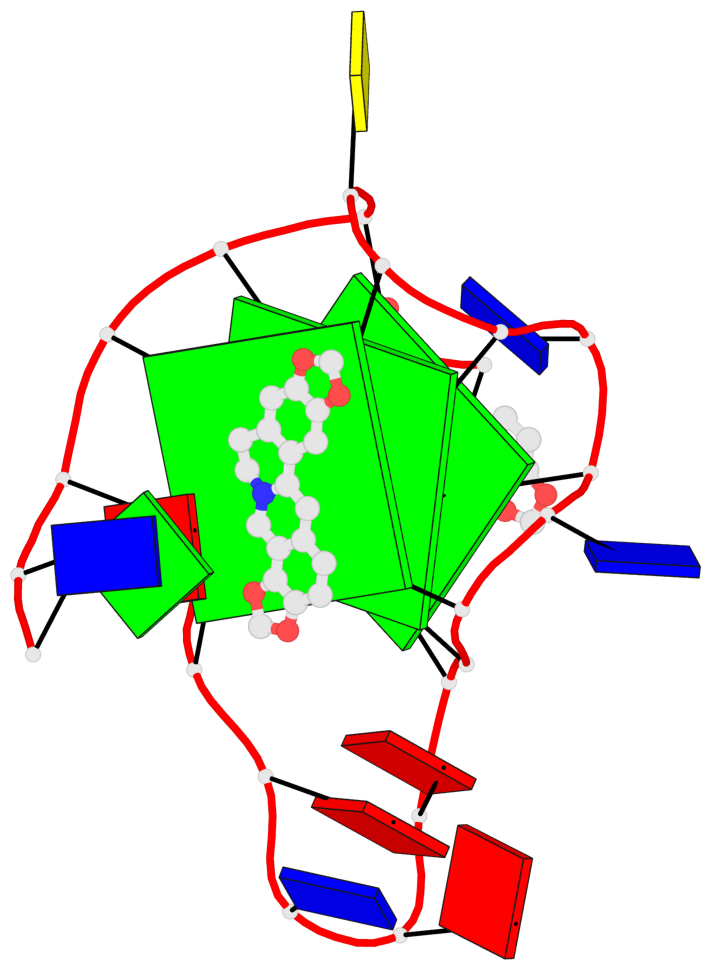
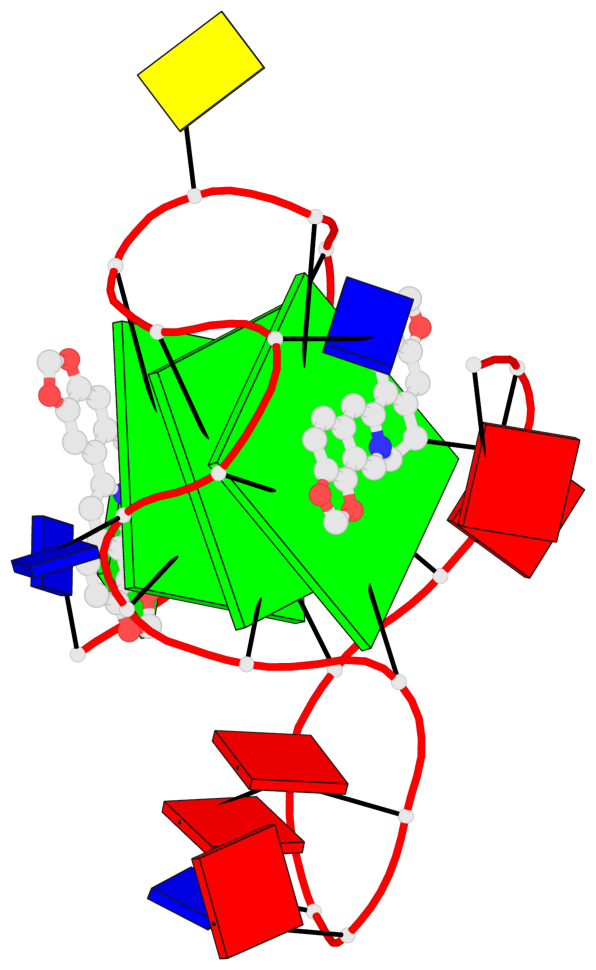
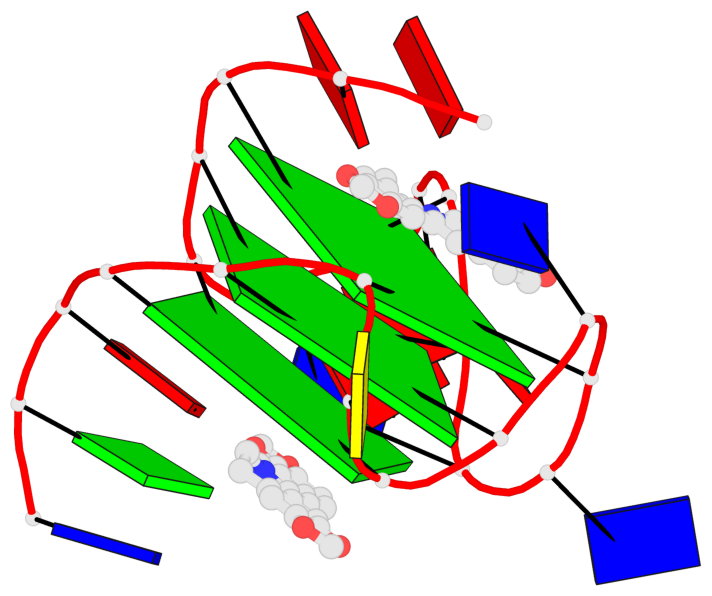
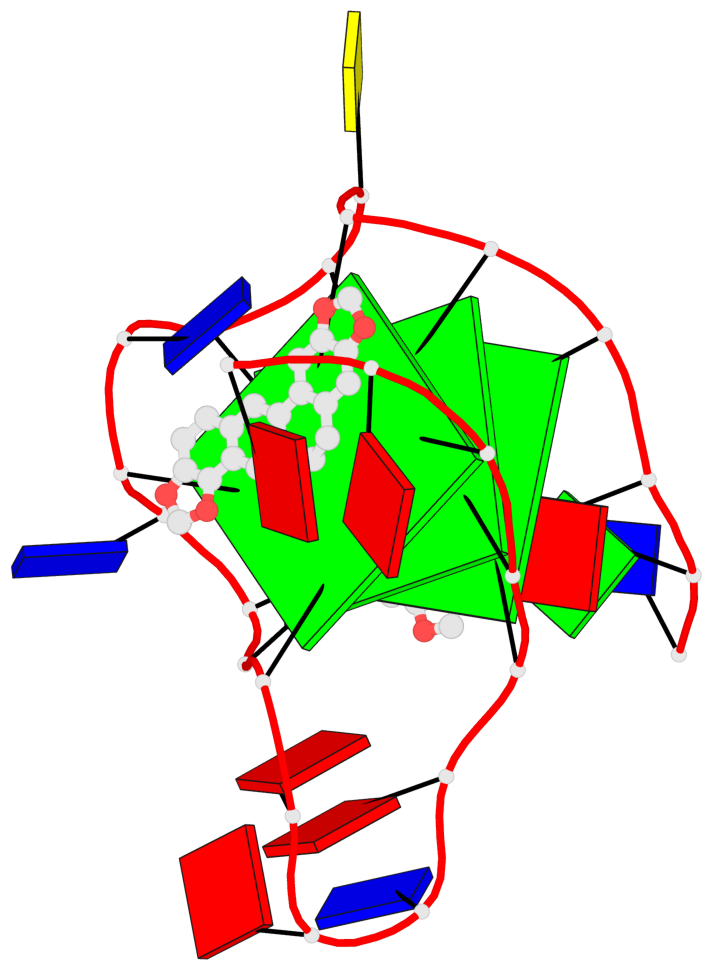
Detailed DSSR results for the G-quadruplex: PDB entry 7x8o
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7x8o
- Class
- DNA
- Method
- NMR
- Summary
- NMR solution structure of the 2:1 coptisine-kras-g4 complex
- Reference
- Wang KB, Liu Y, Li J, Xiao C, Wang Y, Gu W, Li Y, Xia YZ, Yan T, Yang MH, Kong LY (2022): "Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine." Nat Commun, 13, 6016. doi: 10.1038/s41467-022-33761-4.
- Abstract
- KRAS is one of the most highly mutated oncoproteins, which is overexpressed in various human cancers and implicated in poor survival. The G-quadruplex formed in KRAS oncogene promoter (KRAS-G4) is a transcriptional modulator and amenable to small molecule targeting. However, no available KRAS-G4-ligand complex structure has yet been determined, which seriously hinders the structure-based rational design of KRAS-G4 targeting drugs. In this study, we report the NMR solution structures of a bulge-containing KRAS-G4 bound to berberine and coptisine, respectively. The determined complex structure shows a 2:1 binding stoichiometry with each compound recruiting the adjacent flacking adenine residue to form a "quasi-triad plane" that stacks over the two external G-tetrads. The binding involves both π-stacking and electrostatic interactions. Moreover, berberine and coptisine significantly lowered the KRAS mRNA levels in cancer cells. Our study thus provides molecular details of ligand interactions with KRAS-G4 and is beneficial for the design of specific KRAS-G4-interactive drugs.
- G4 notes
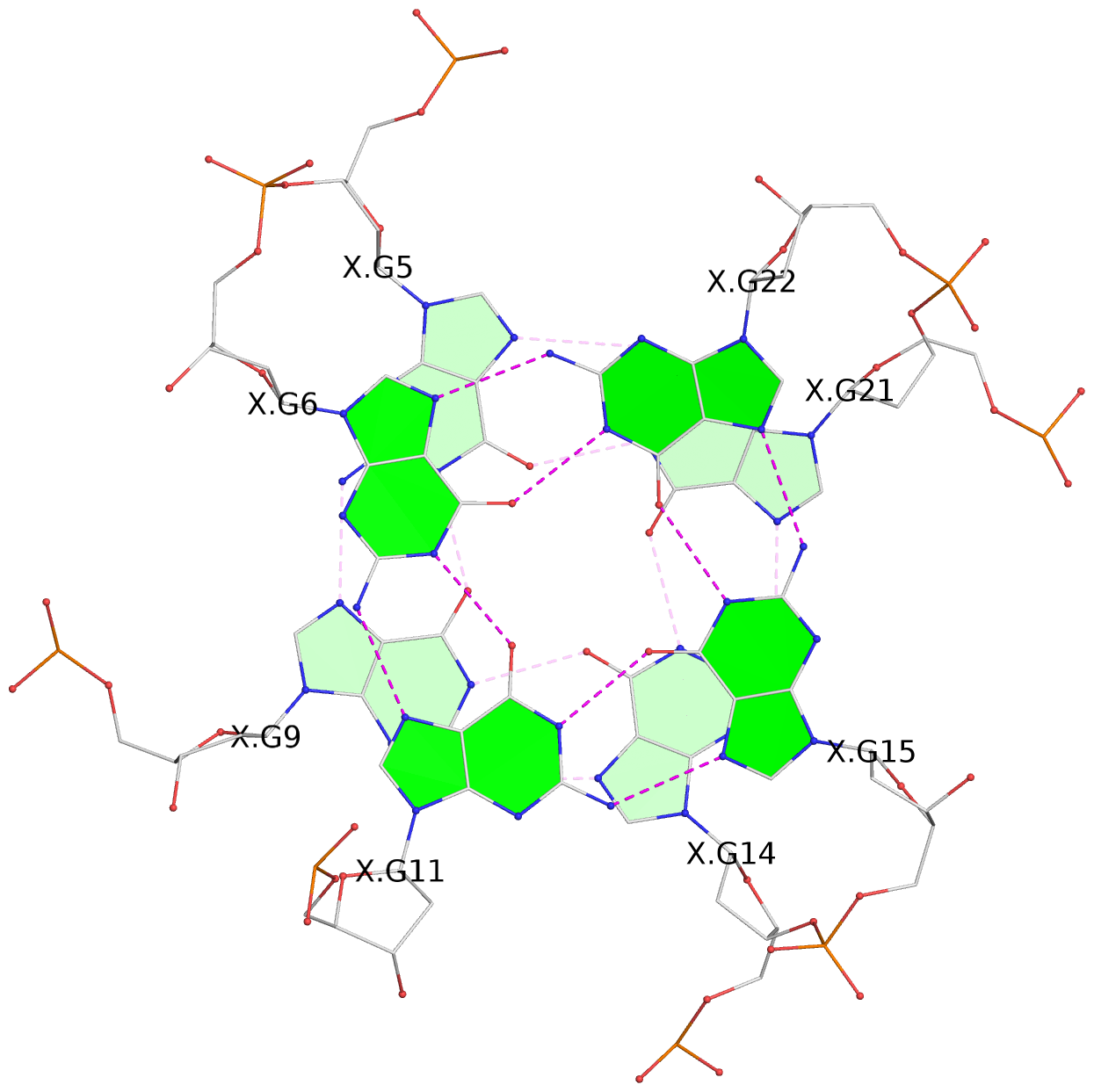
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
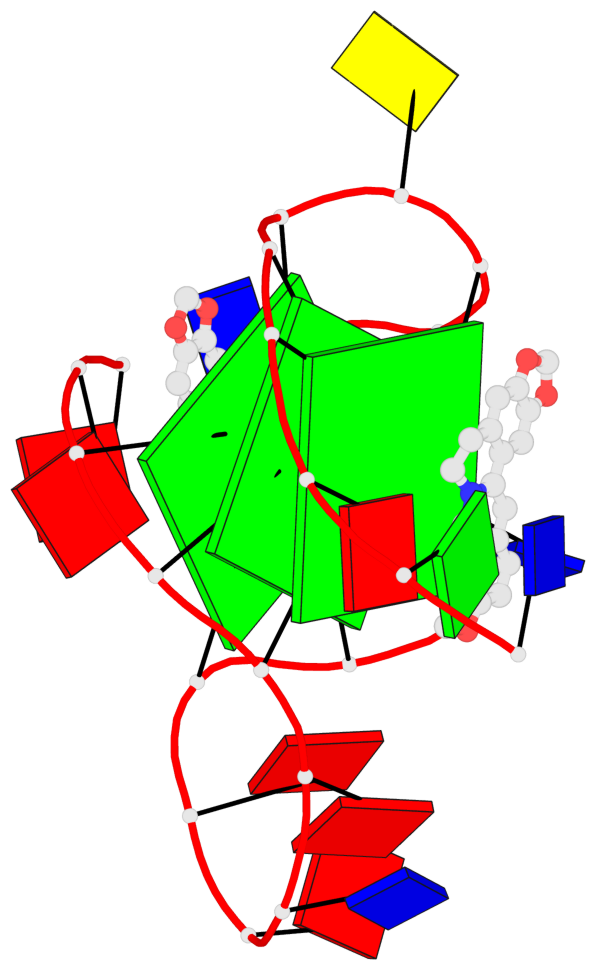
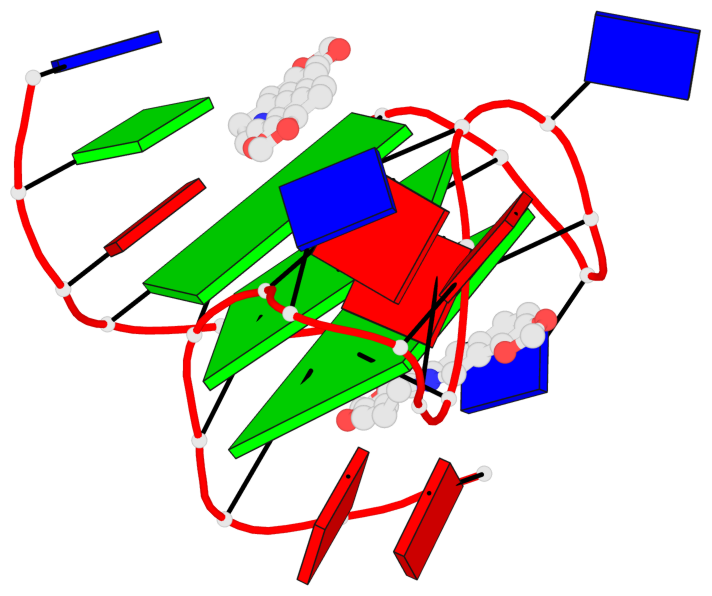
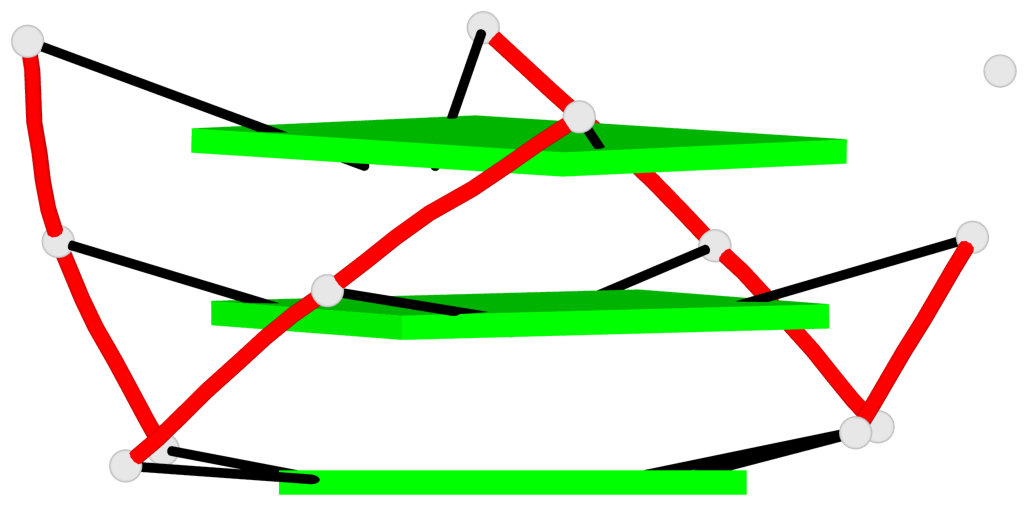
Base-block schematics in six views
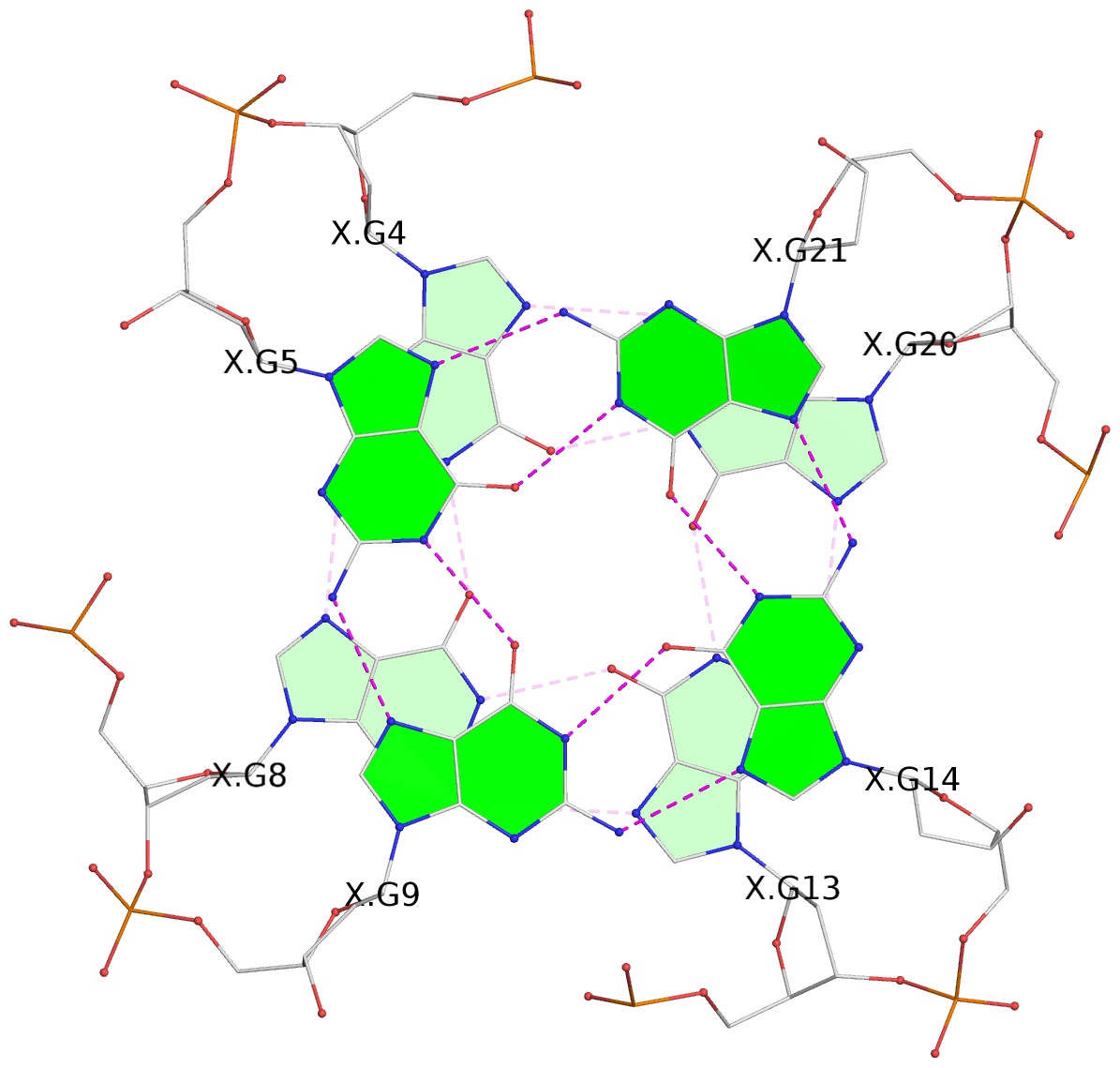
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.101 type=planar nts=4 GGGG X.DG4,X.DG8,X.DG13,X.DG20 2 glyco-bond=---- sugar=-.-- groove=---- planarity=0.238 type=other nts=4 GGGG X.DG5,X.DG9,X.DG14,X.DG21 3 glyco-bond=---- sugar=---- groove=---- planarity=0.301 type=bowl nts=4 GGGG X.DG6,X.DG11,X.DG15,X.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.