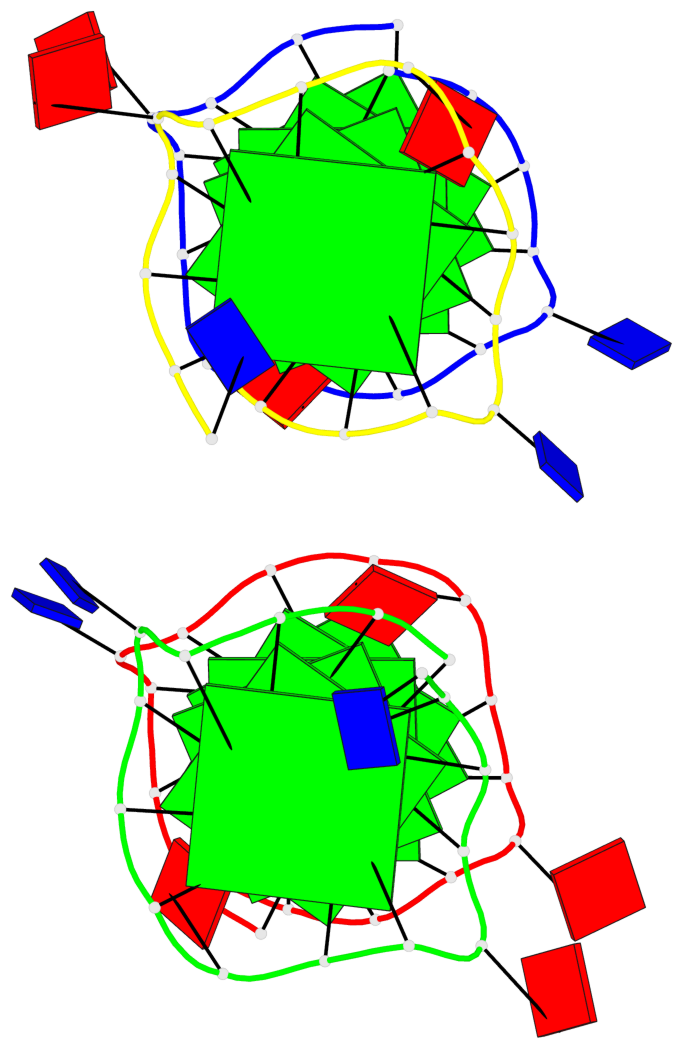
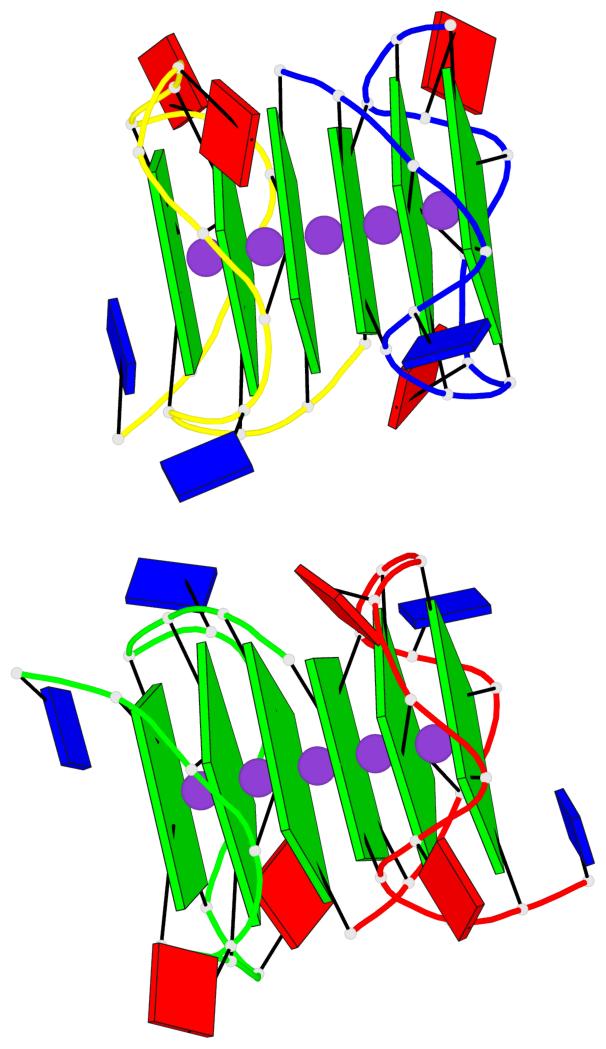
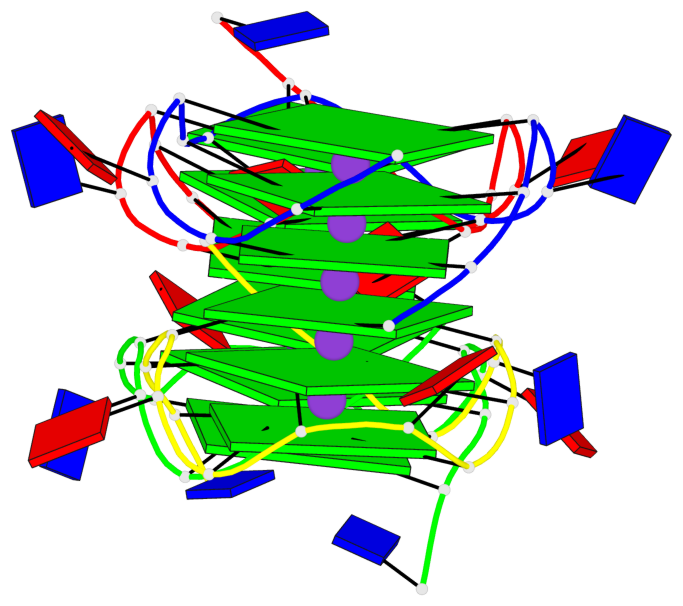
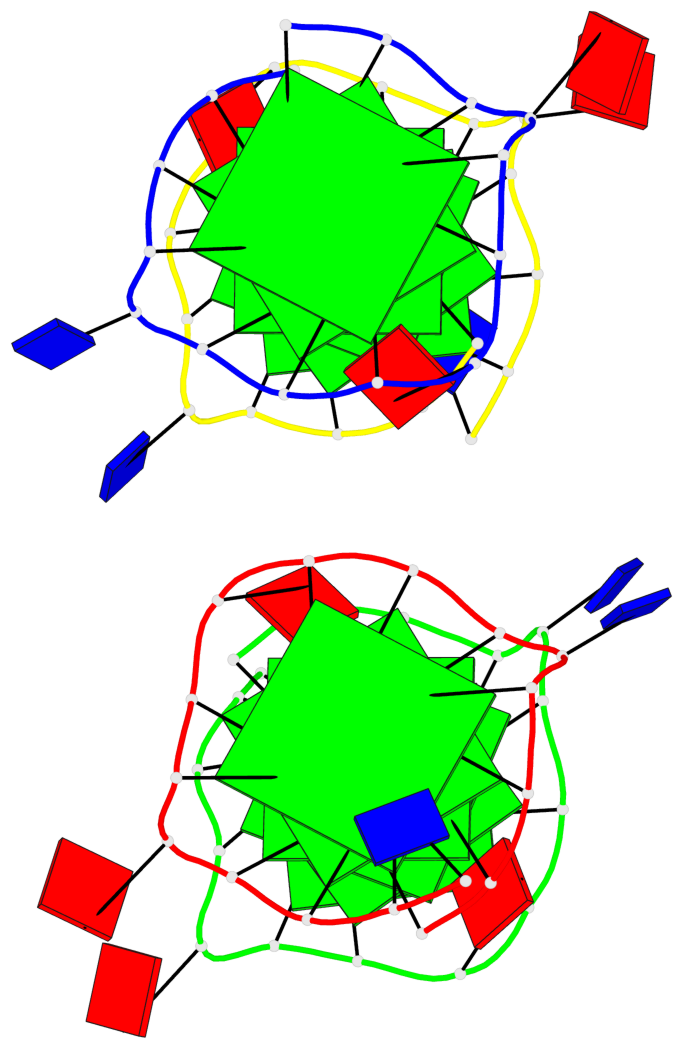
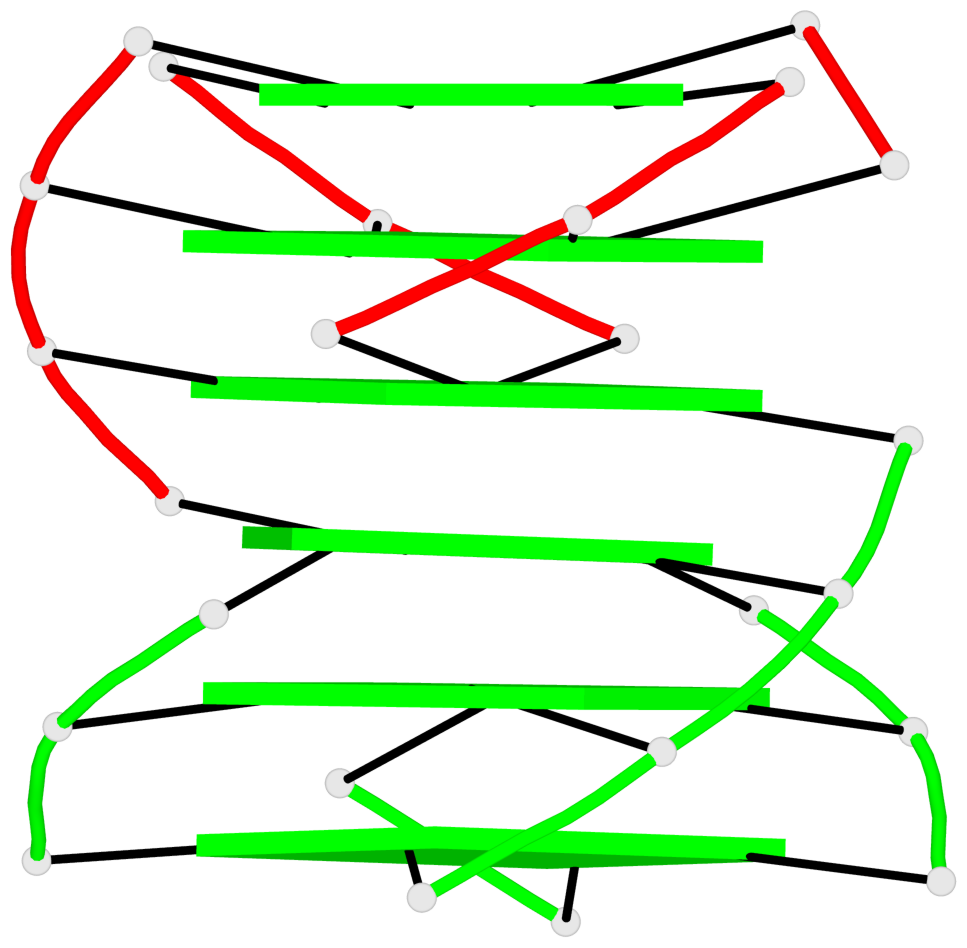
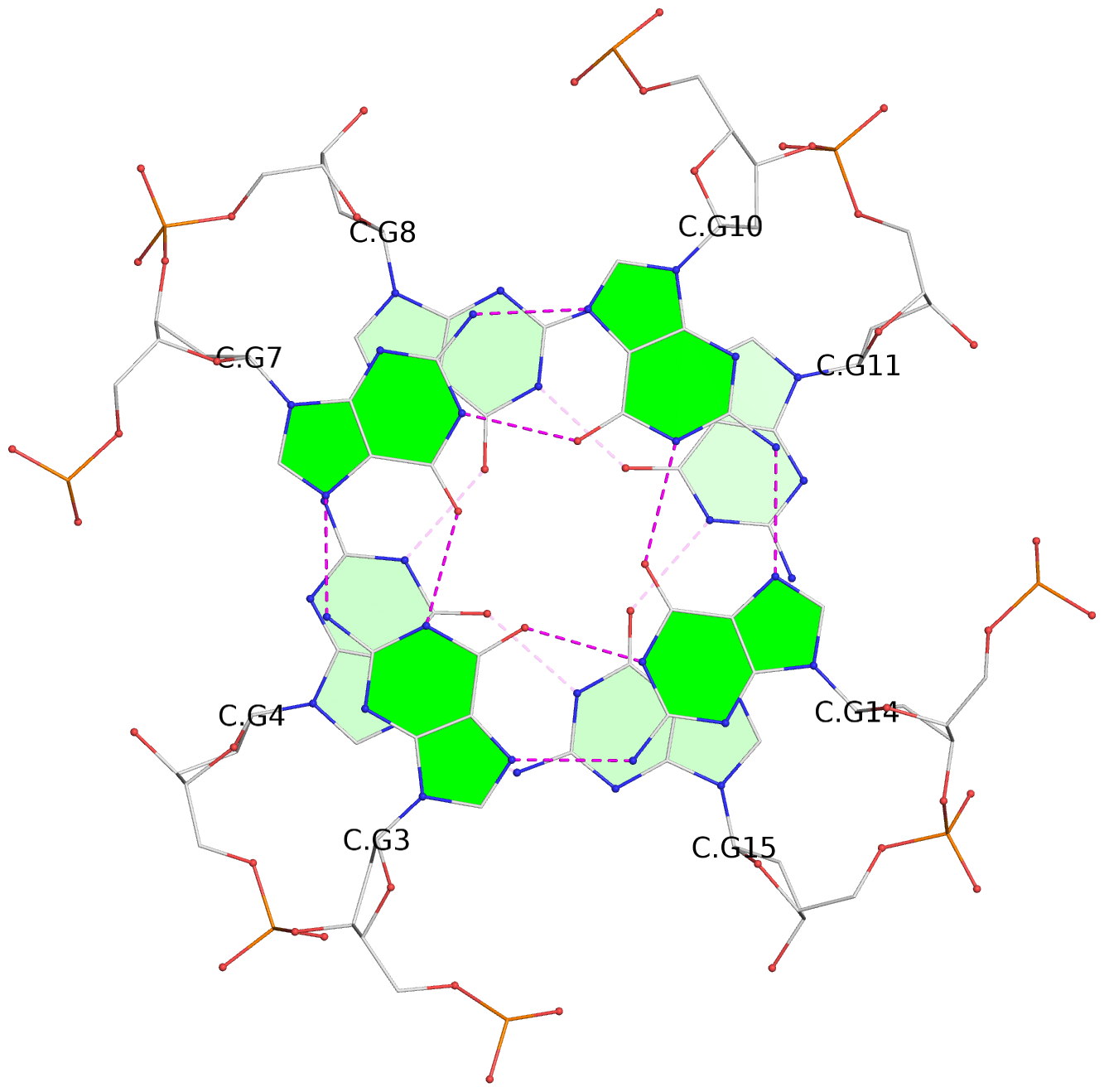
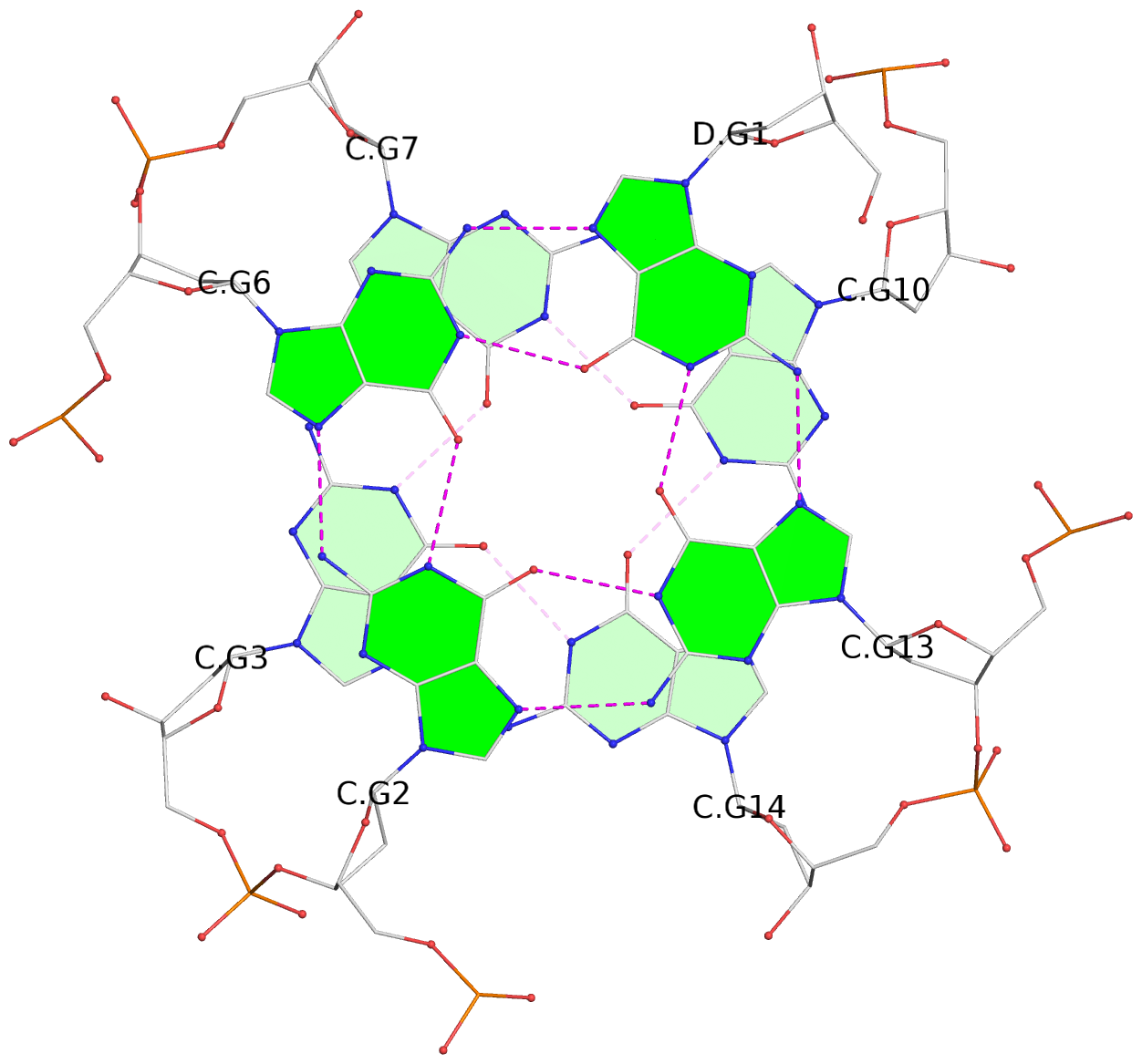
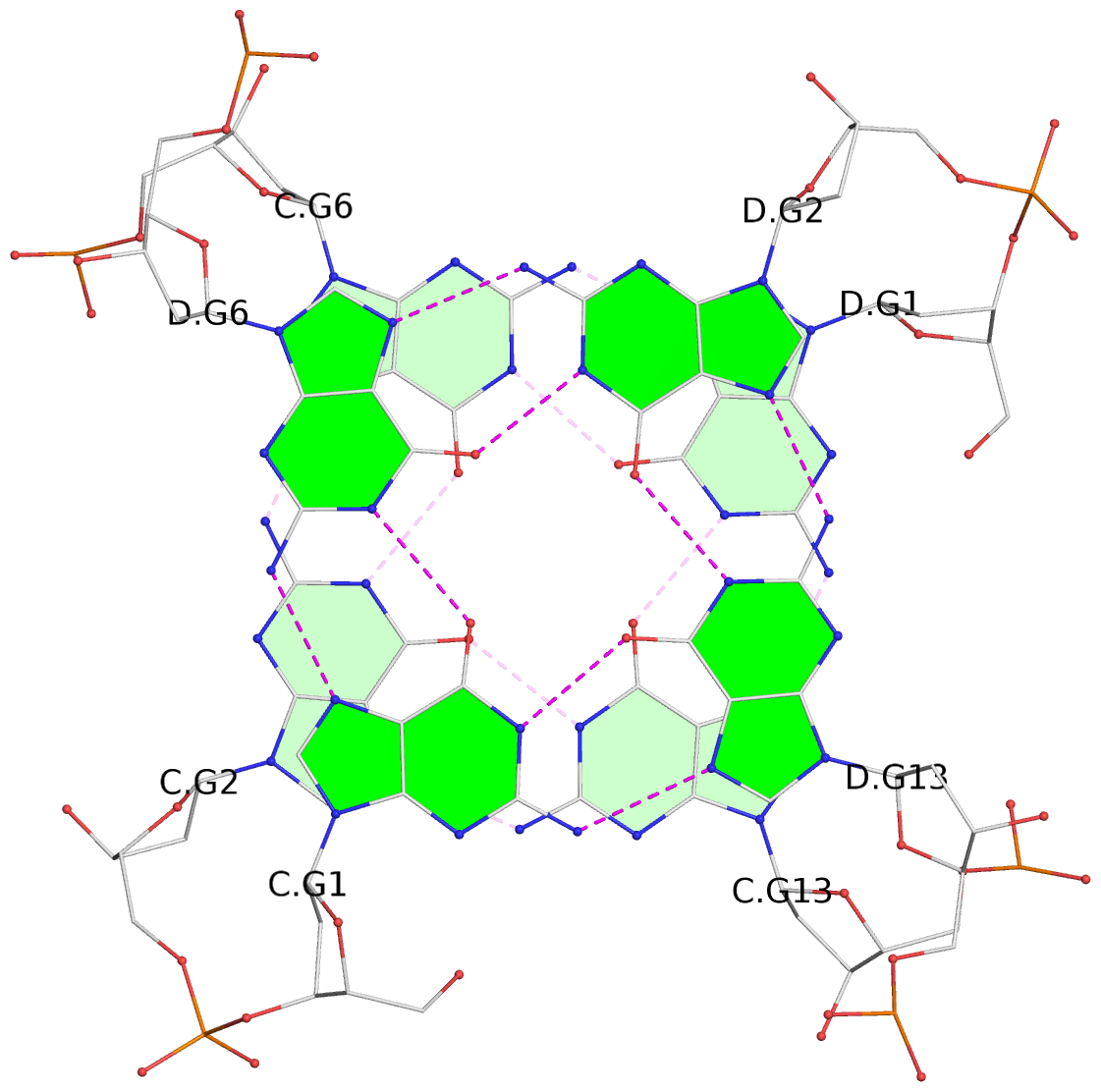
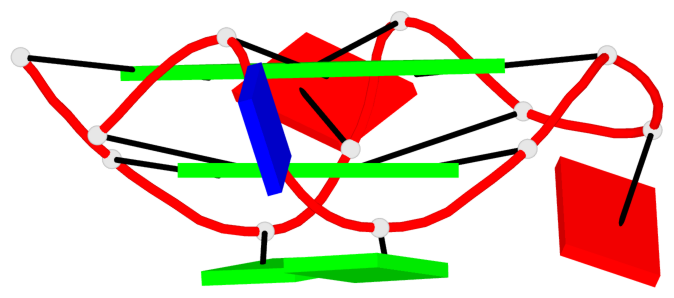
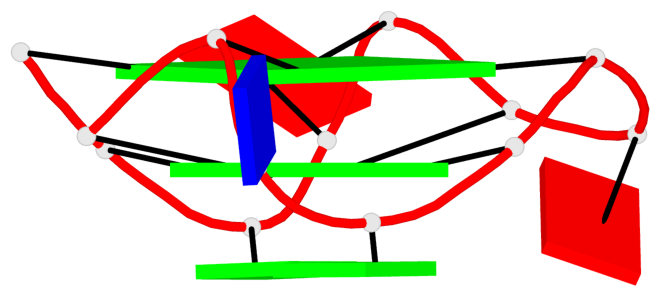
Detailed DSSR results for the G-quadruplex: PDB entry 7xdh
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7xdh
- Class
- DNA
- Method
- X-ray (1.83 Å)
- Summary
- Crystal structure of a dimeric interlocked parallel g-quadruplex
- Reference
- Ngo KH, Liew CW, Lattmann S, Winnerdy FR, Phan AT (2022): "Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex." Biochem.Biophys.Res.Commun., 613, 153-158. doi: 10.1016/j.bbrc.2022.04.020.
- Abstract
- 93del is a 16-nucleotide G-quadruplex-forming aptamer which can inhibit the activity of the HIV-1 integrase enzyme at nanomolar concentration. Previous structural analyses of 93del using NMR spectroscopy have shown that the aptamer forms an interlocked G-quadruplex structure in K+ solution. Due to its exceptional stability and unique topology, 93del has been used in many different studies involving DNA G-quadruplexes, such as DNA aptamer and multimer design, as well as DNA fluorescence research. To gain further insights on the structure of this unique aptamer, we have determined several high-resolution crystal structures of 93del and its variants. While confirming the overall dimeric interlocked G-quadruplex folding topology previously determined by NMR, our results reveal important detailed structural information, particularly the formation of a water-mediated A•G•G•G•G pentad. These insights allow us to better understand the formation of various structural elements in G-quadruplexes and should be useful for designing and manipulating G-quadruplex scaffolds with desired properties.
- G4 notes
- 12 G-tetrads, 2 G4 helices, 4 G4 stems, 2 G4 coaxial stacks, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'-SEPARATED
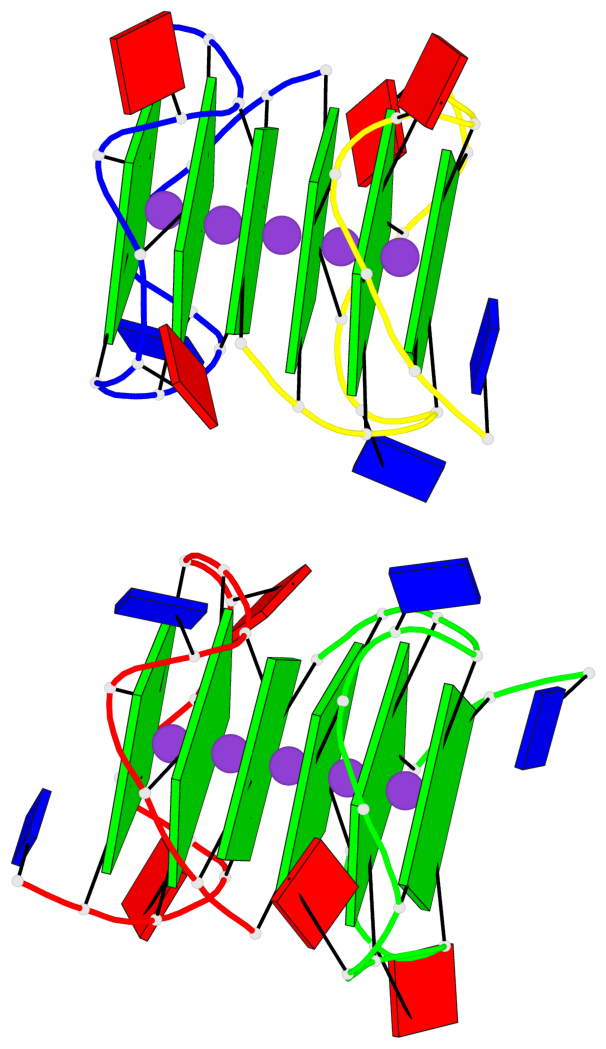
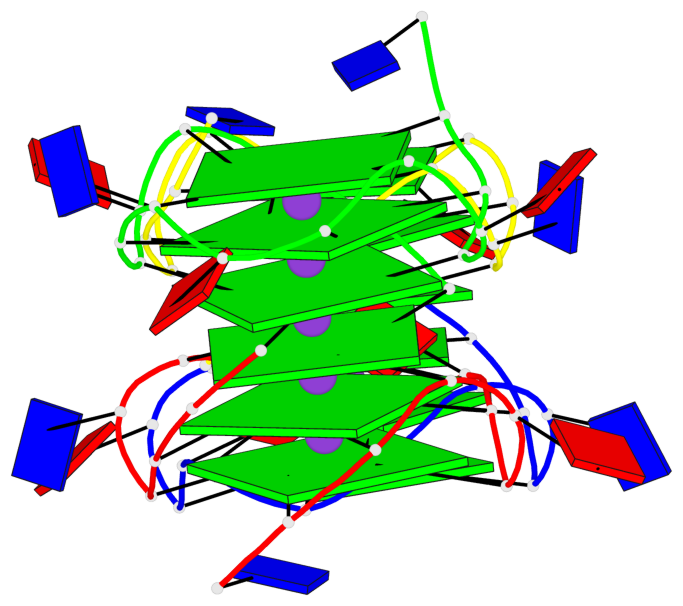
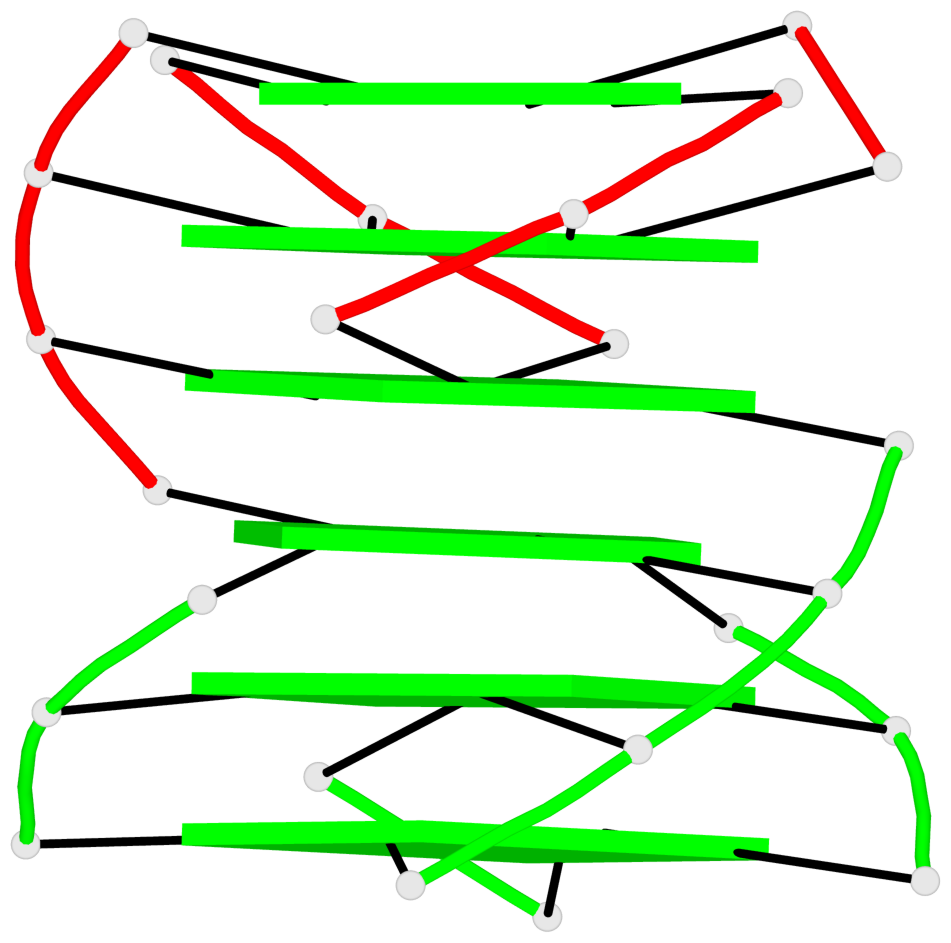
Base-block schematics in six views
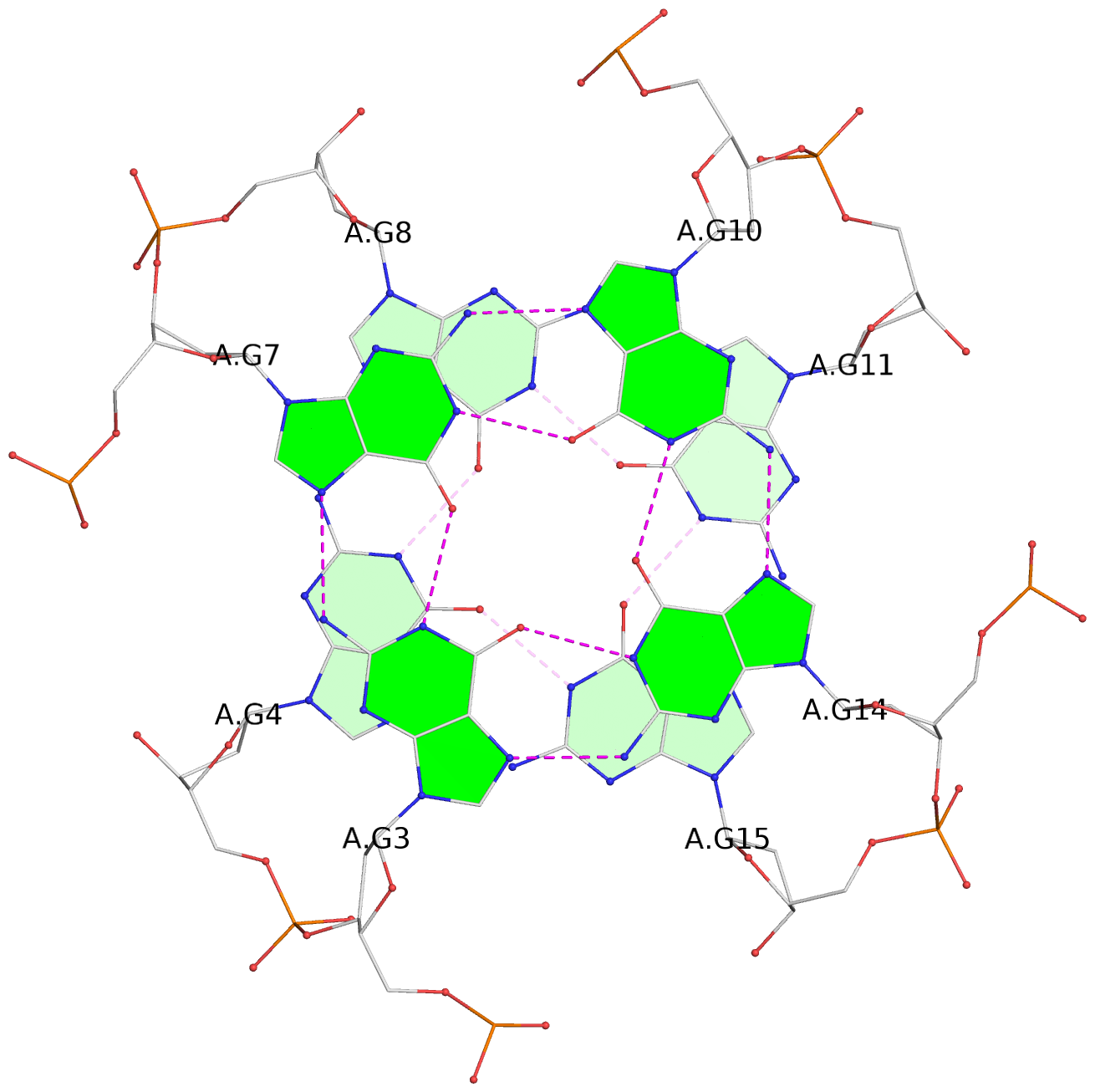
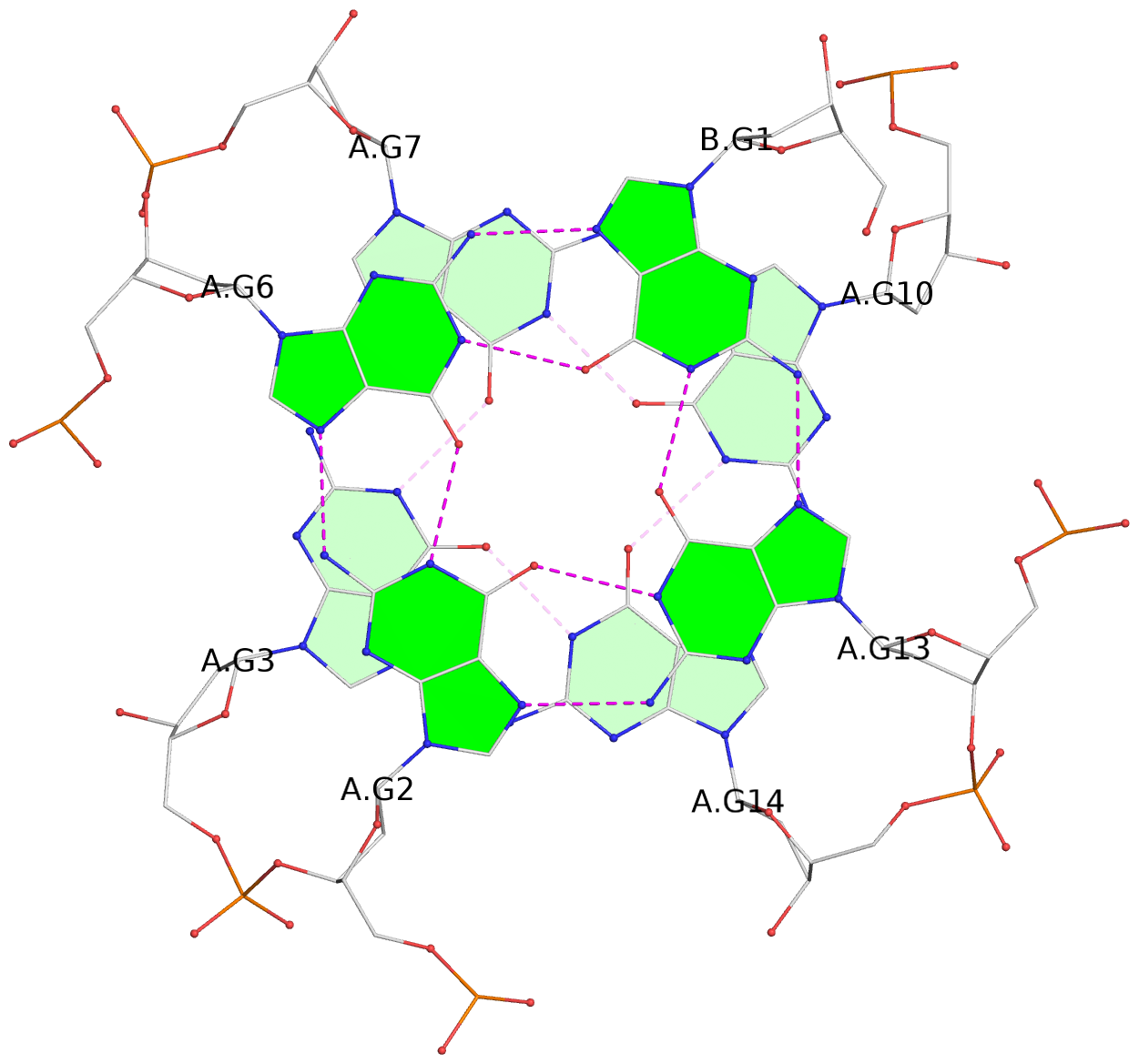
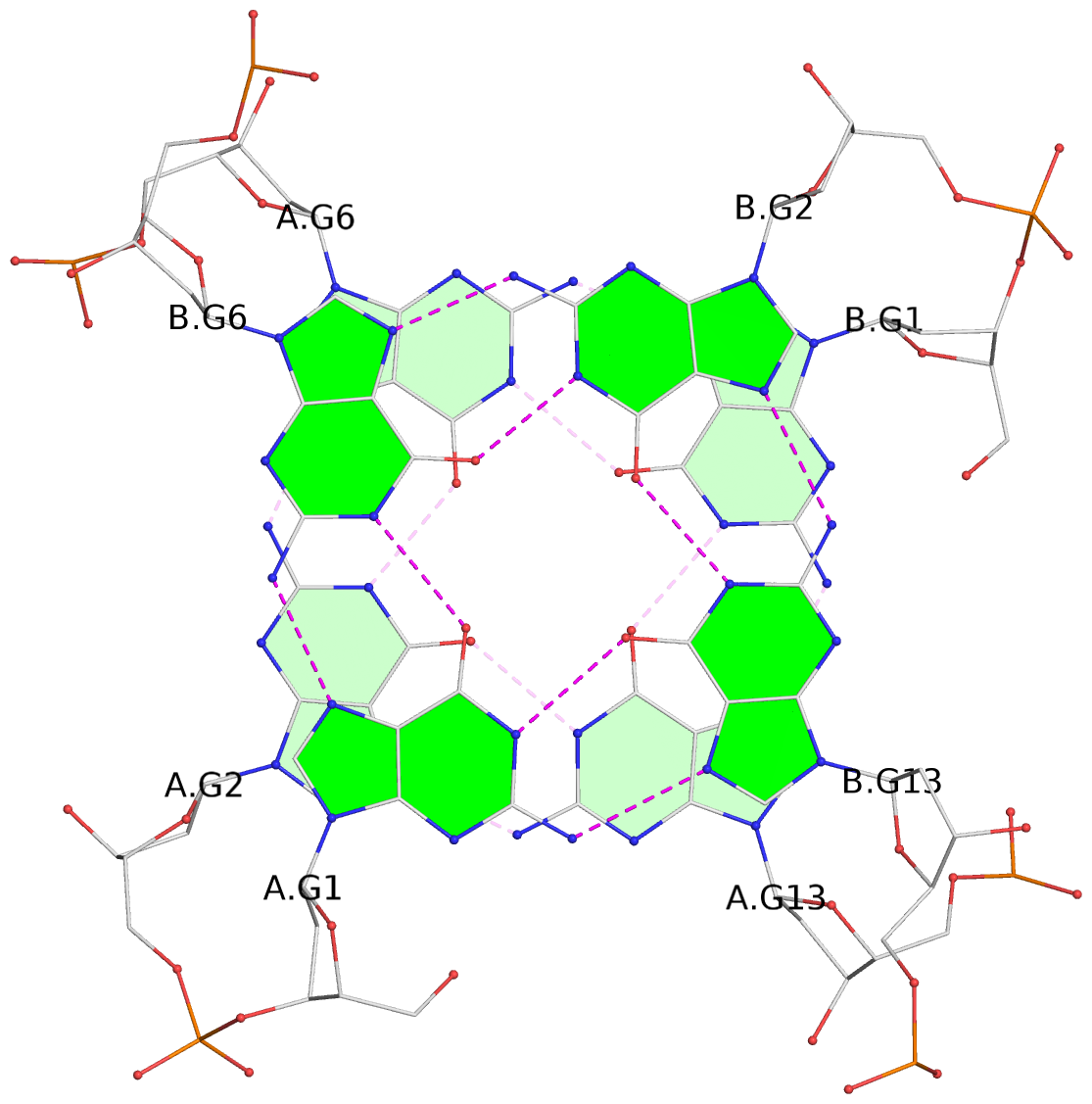
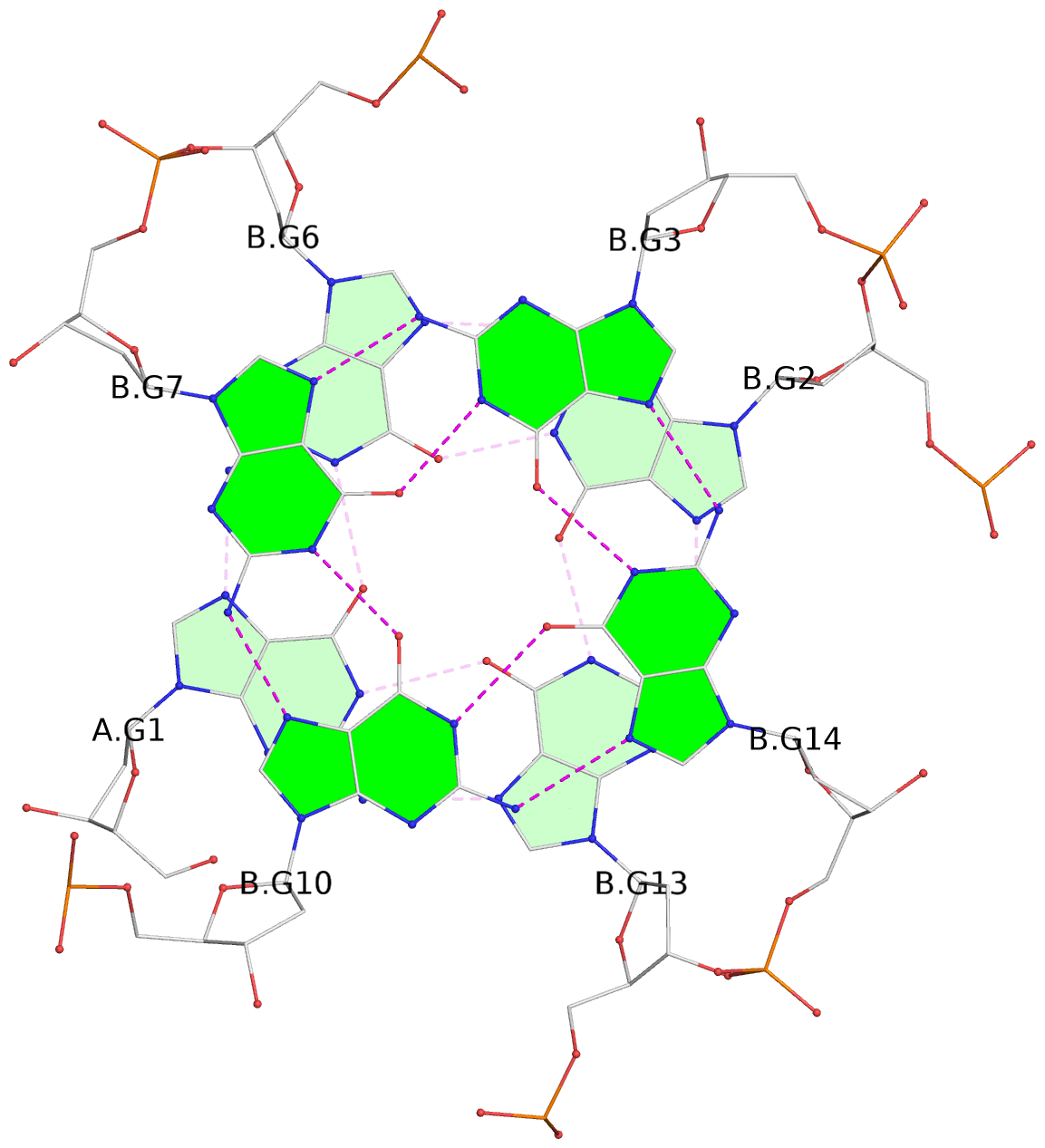
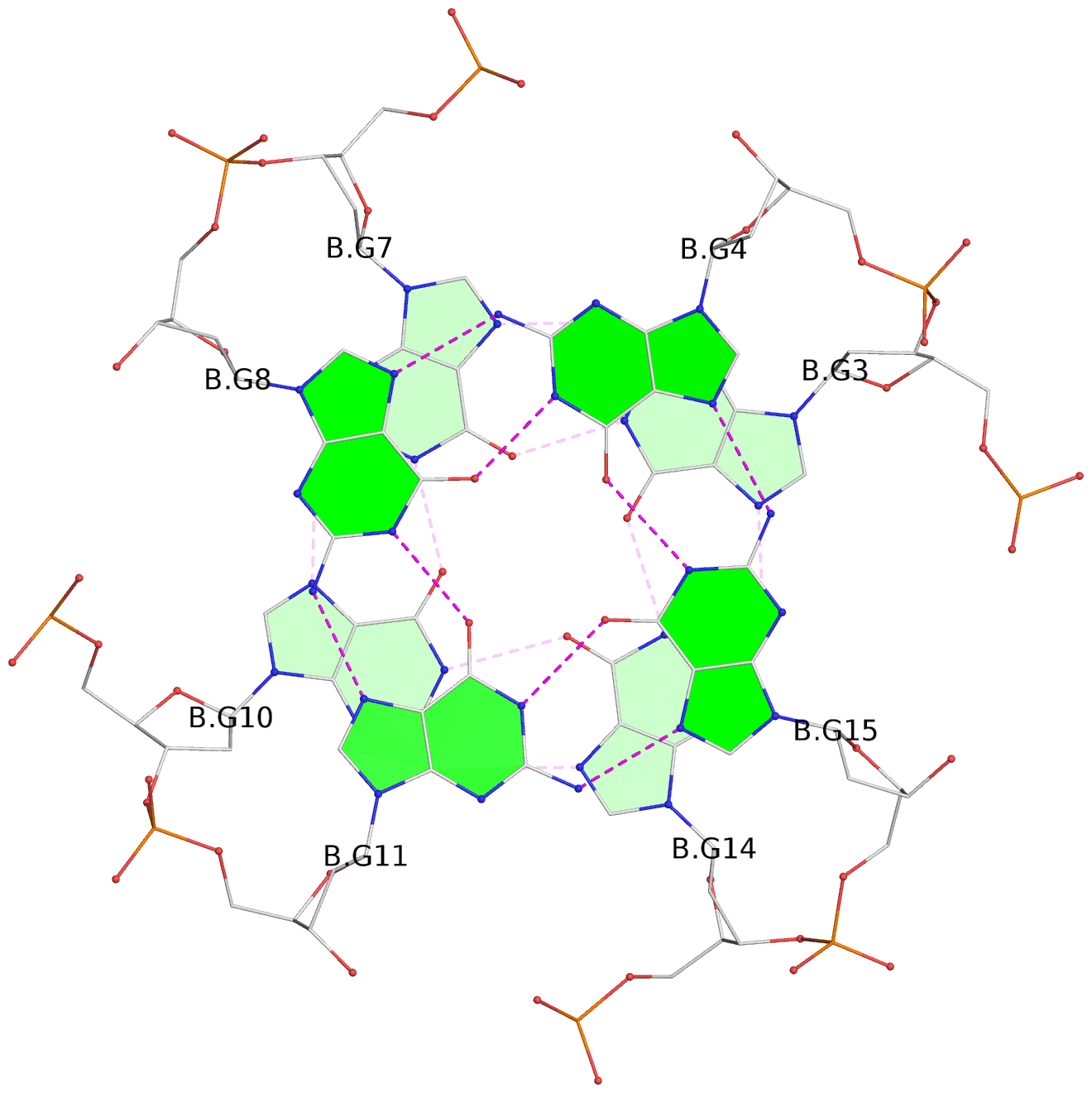
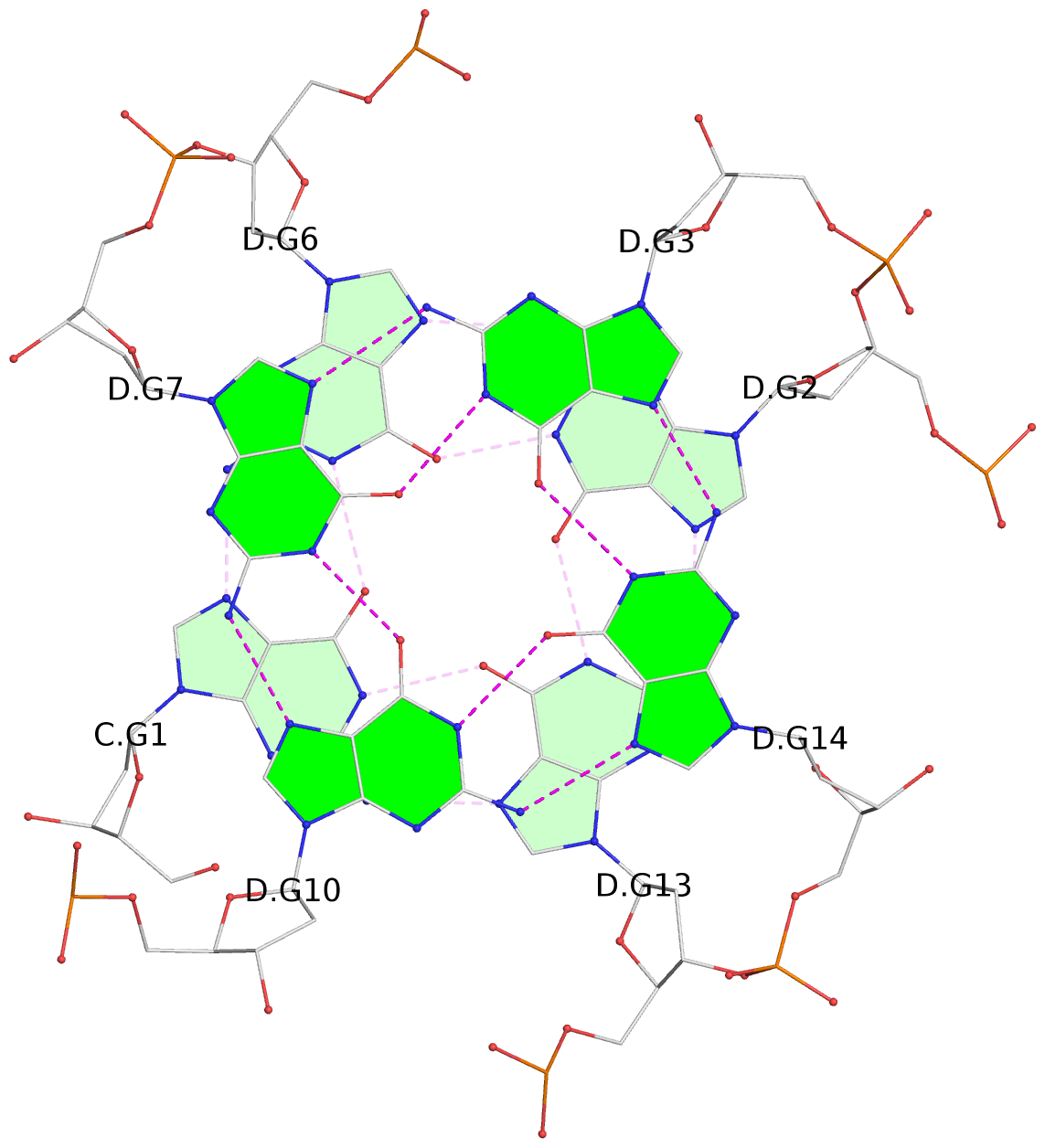
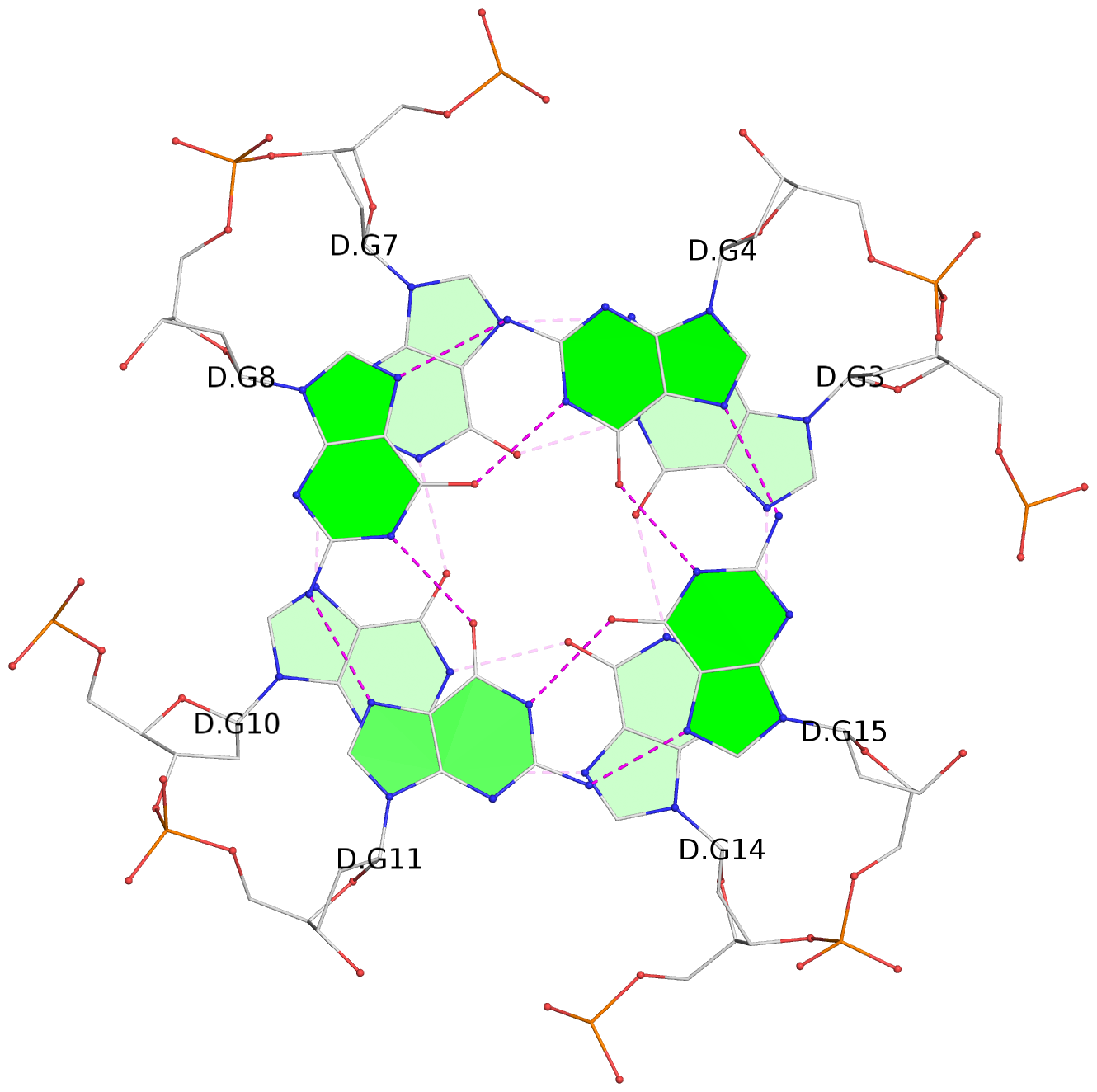
List of 12 G-tetrads
1 glyco-bond=s--- sugar=---. groove=w--n planarity=0.093 type=planar nts=4 GGGG A.DG1,B.DG6,B.DG2,B.DG13 2 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.127 type=planar nts=4 GGGG A.DG2,A.DG6,B.DG1,A.DG13 3 glyco-bond=---- sugar=.-.- groove=---- planarity=0.086 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG10,A.DG14 4 glyco-bond=---- sugar=---- groove=---- planarity=0.238 type=other nts=4 GGGG A.DG4,A.DG8,A.DG11,A.DG15 5 glyco-bond=---- sugar=.-.- groove=---- planarity=0.082 type=planar nts=4 GGGG B.DG3,B.DG7,B.DG10,B.DG14 6 glyco-bond=---- sugar=---- groove=---- planarity=0.208 type=other nts=4 GGGG B.DG4,B.DG8,B.DG11,B.DG15 7 glyco-bond=s--- sugar=-.-. groove=w--n planarity=0.117 type=planar nts=4 GGGG C.DG1,D.DG6,D.DG2,D.DG13 8 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.120 type=planar nts=4 GGGG C.DG2,C.DG6,D.DG1,C.DG13 9 glyco-bond=---- sugar=.-.- groove=---- planarity=0.103 type=planar nts=4 GGGG C.DG3,C.DG7,C.DG10,C.DG14 10 glyco-bond=---- sugar=---- groove=---- planarity=0.231 type=other nts=4 GGGG C.DG4,C.DG8,C.DG11,C.DG15 11 glyco-bond=---- sugar=.-.- groove=---- planarity=0.085 type=planar nts=4 GGGG D.DG3,D.DG7,D.DG10,D.DG14 12 glyco-bond=---- sugar=---- groove=---- planarity=0.223 type=other nts=4 GGGG D.DG4,D.DG8,D.DG11,D.DG15
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
Helix#2, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#3, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#4, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 2 G4 coaxial stacks
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5'-SEPARATED] 2 G4 helix#2 contains 2 G4 stems: [#3,#4] [5'/5'-SEPARATED]