Detailed DSSR results for the G-quadruplex: PDB entry 7zem
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7zem
- Class
- DNA
- Method
- NMR
- Summary
- Structure of a parallel g-quadruplex with a snapback loop
- Reference
- Jana J, Vianney YM, Schroder N, Weisz K (2022): "Guiding the folding of G-quadruplexes through loop residue interactions." Nucleic Acids Res., 50, 7161-7175. doi: 10.1093/nar/gkac549.
- Abstract
- A G-rich sequence was designed to allow folding into either a stable parallel or hybrid-type topology. With the parent sequence featuring coexisting species, various related sequences with single and double mutations and with a shortened central propeller loop affected the topological equilibrium. Two simple modifications, likewise introduced separately to all sequences, were employed to lock folds into one of the topologies without noticeable structural alterations. The unique combination of sequence mutations, high-resolution NMR structural information, and the thermodynamic stability for both topological competitors identified critical loop residue interactions. In contrast to first loop residues, which are mostly disordered and exposed to solvent in both propeller and lateral loops bridging a narrow groove, the last loop residue in a lateral three-nucleotide loop is engaged in stabilizing stacking interactions. The propensity of single-nucleotide loops to favor all-parallel topologies by enforcing a propeller-like conformation of an additional longer loop is shown to result from their preference in linking two outer tetrads of the same tetrad polarity. Taken together, the present studies contribute to a better structural and thermodynamic understanding of delicate loop interactions in genomic and artificially designed quadruplexes, e.g. when employed as therapeutics or in other biotechnological applications.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
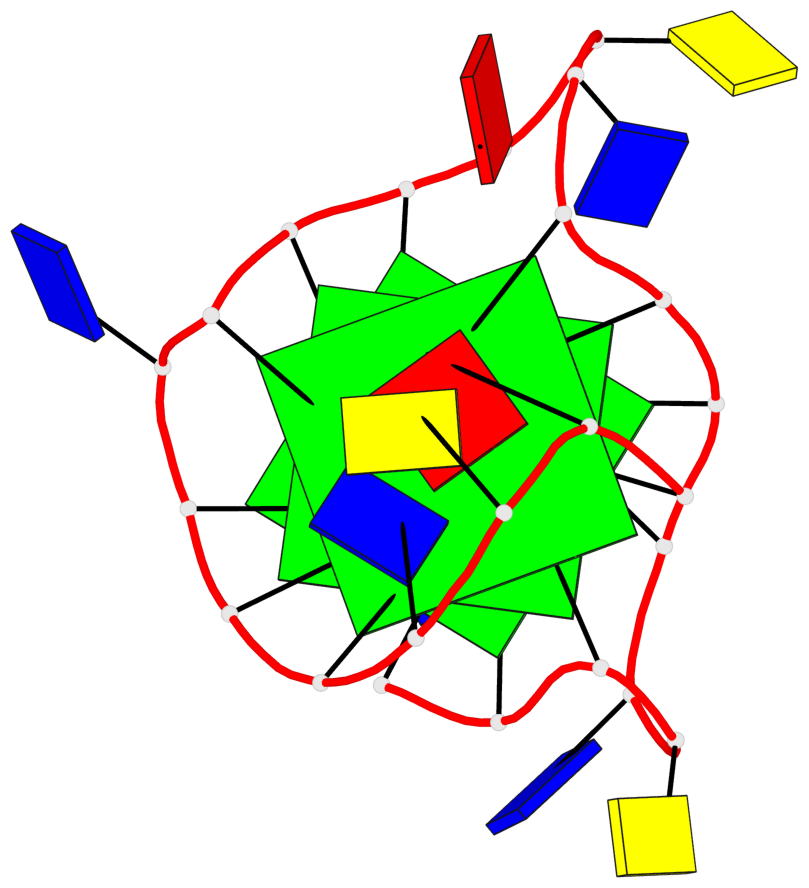
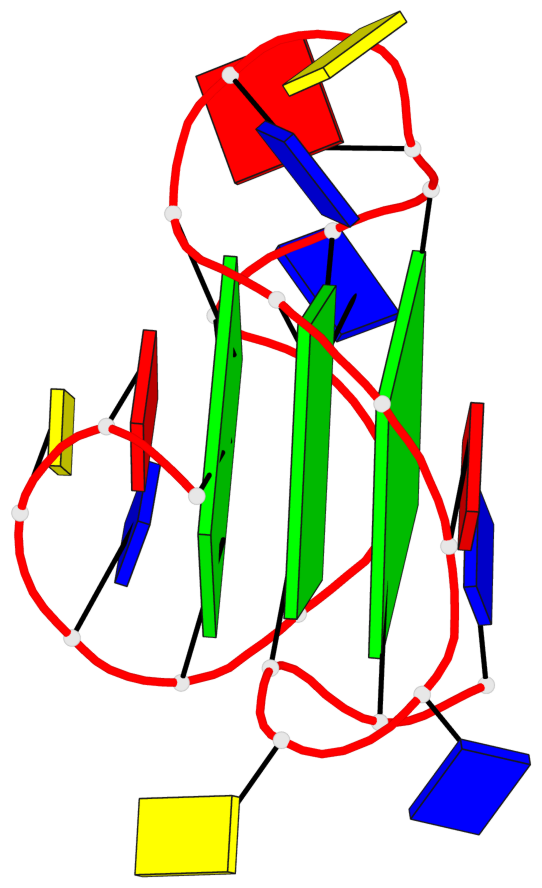
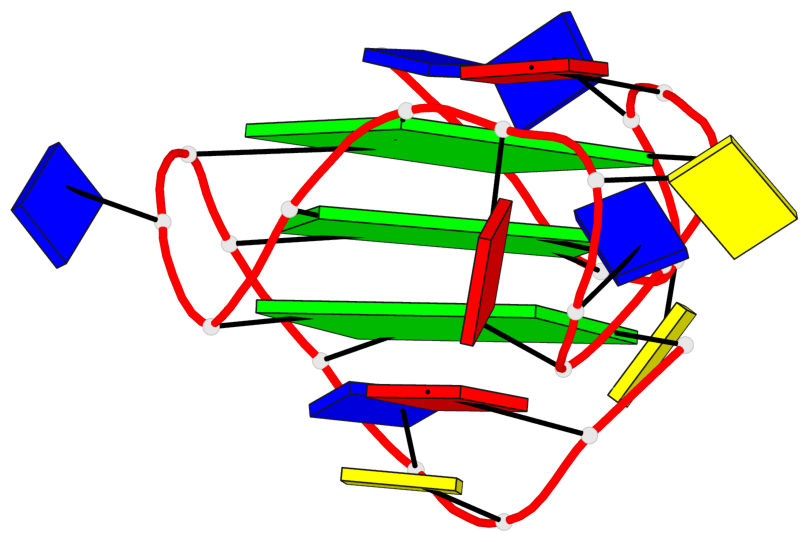
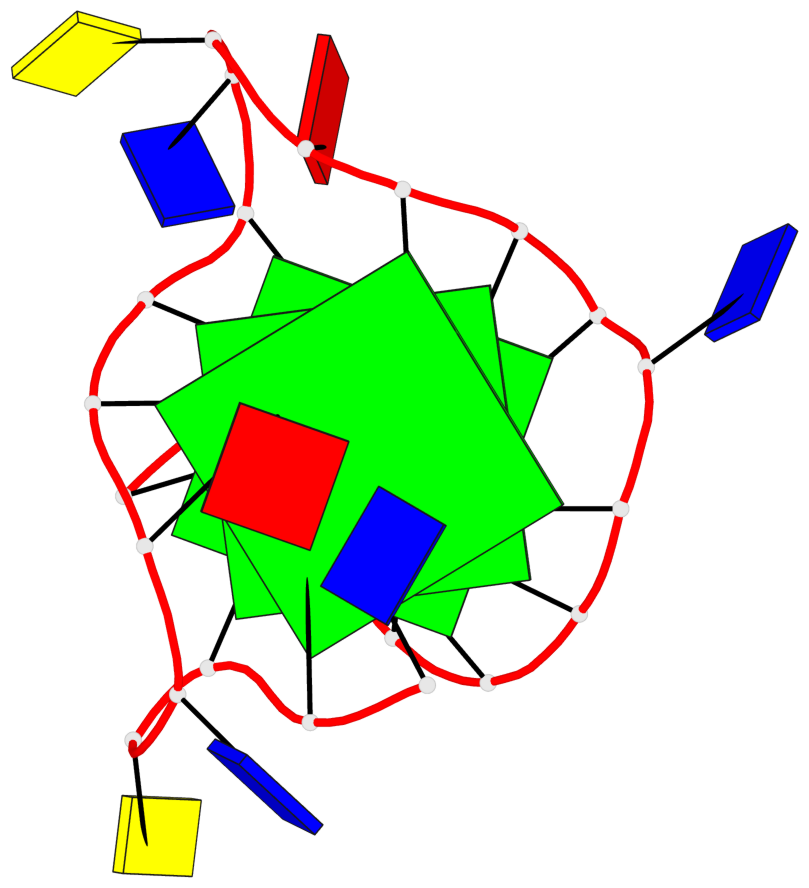
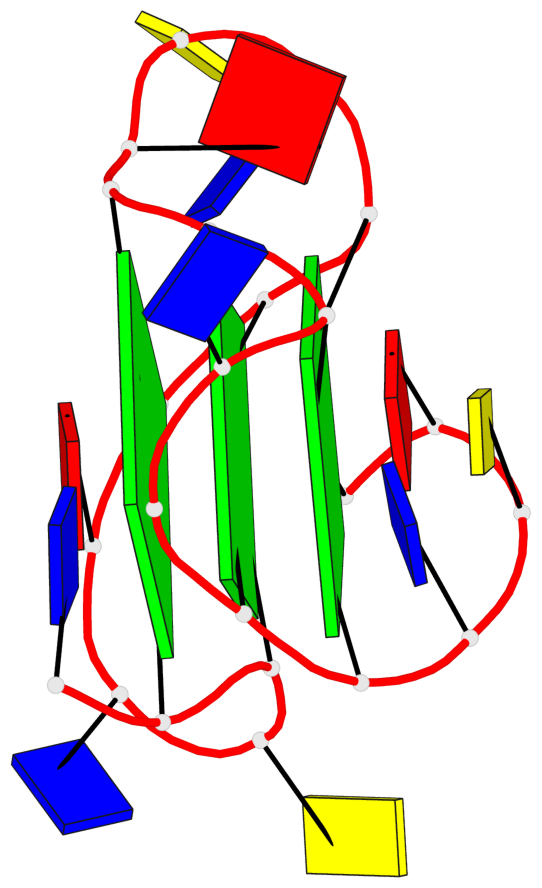
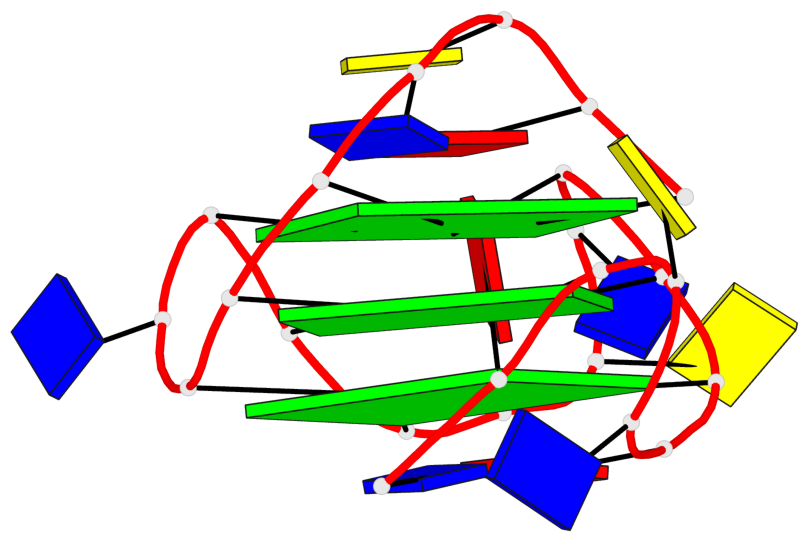
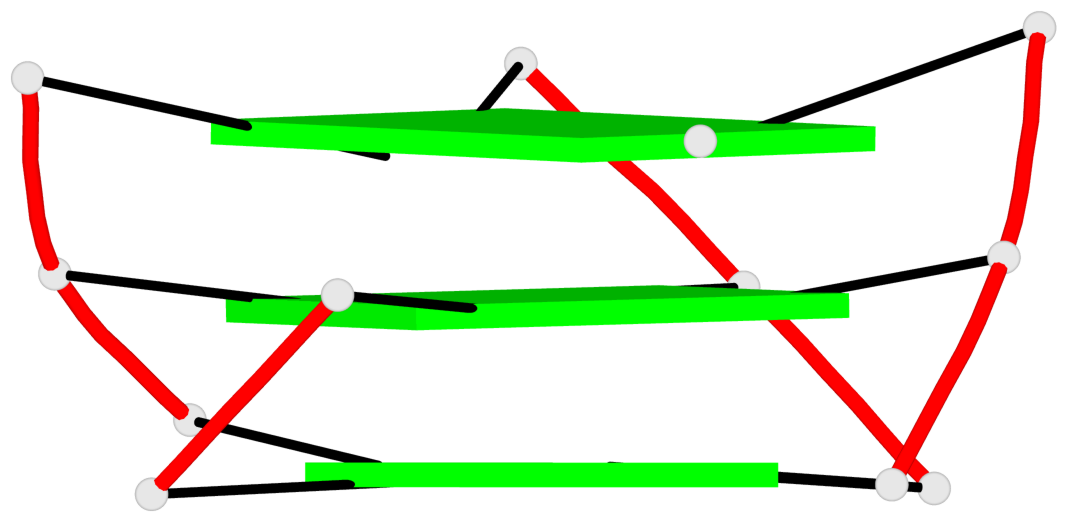
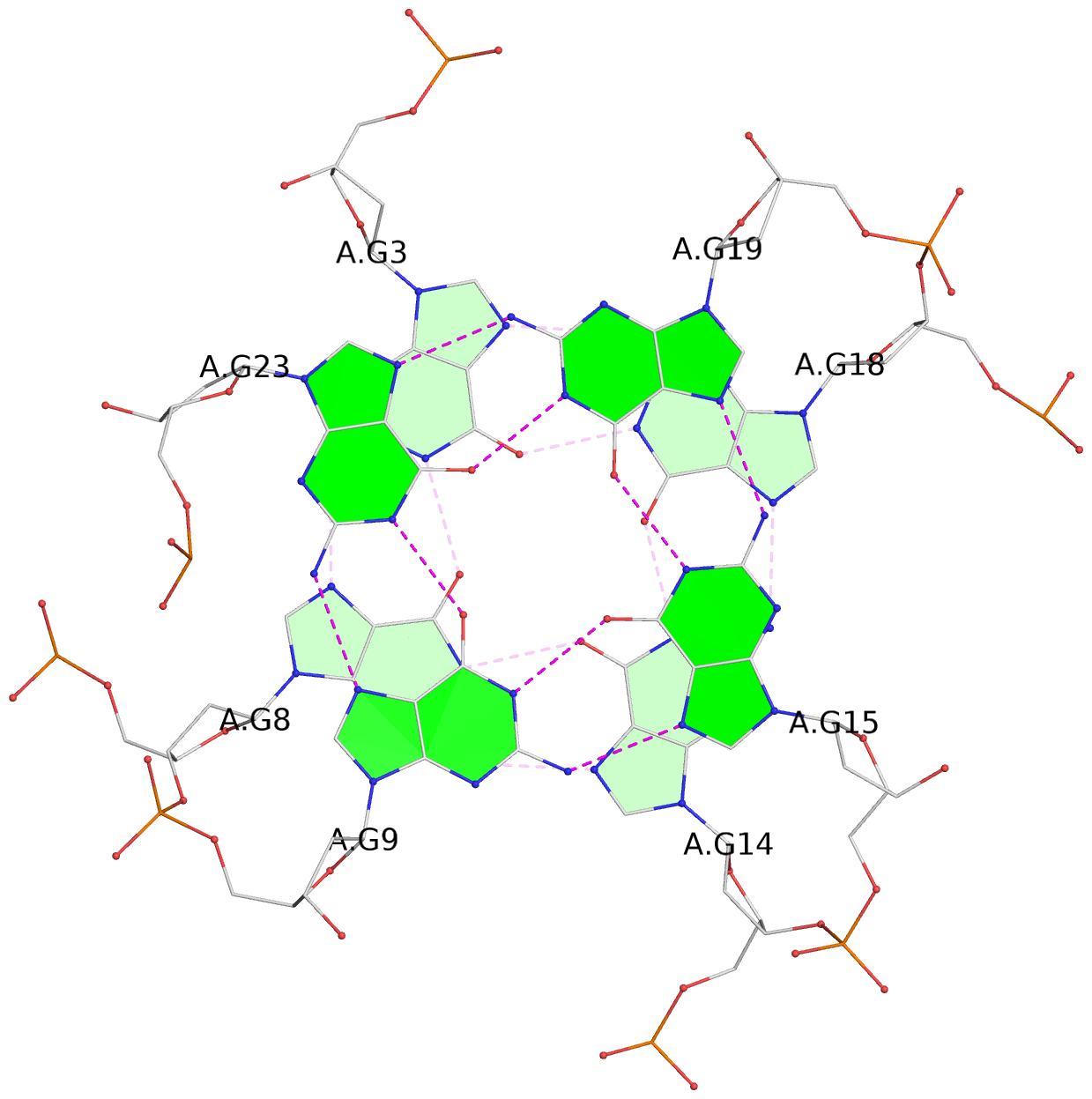
Base-block schematics in six views
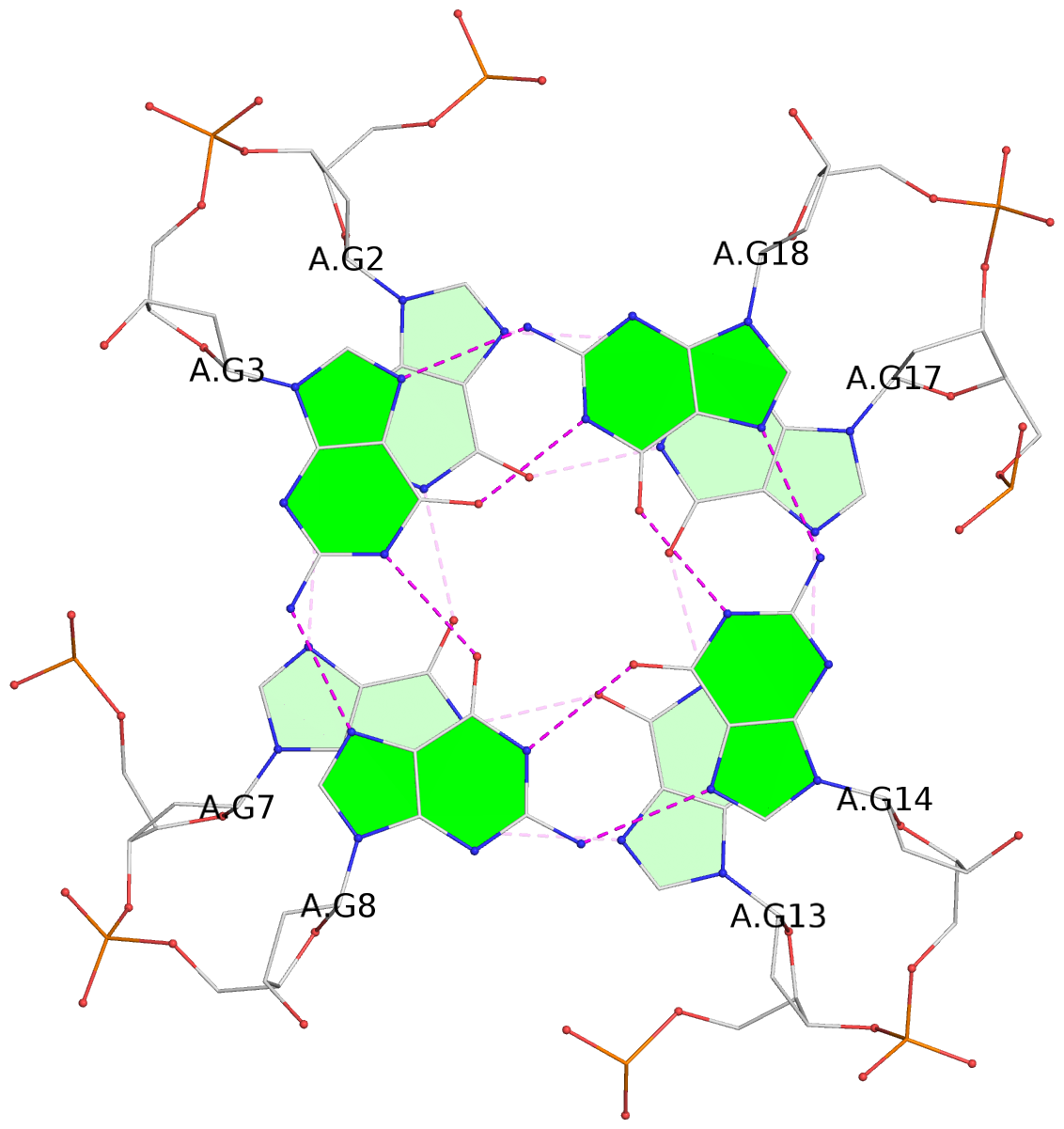
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.150 type=planar nts=4 GGGG A.DG2,A.DG7,A.DG13,A.DG17 2 glyco-bond=---- sugar=---- groove=---- planarity=0.137 type=planar nts=4 GGGG A.DG3,A.DG8,A.DG14,A.DG18 3 glyco-bond=---s sugar=---. groove=--wn planarity=0.264 type=bowl nts=4 GGGG A.DG9,A.DG15,A.DG19,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 non-stem G4-loop (including the two closing Gs)
1 type=lateral helix=#1 nts=5 GTCAG A.DG19,A.DT20,A.DC21,A.DA22,A.DG23