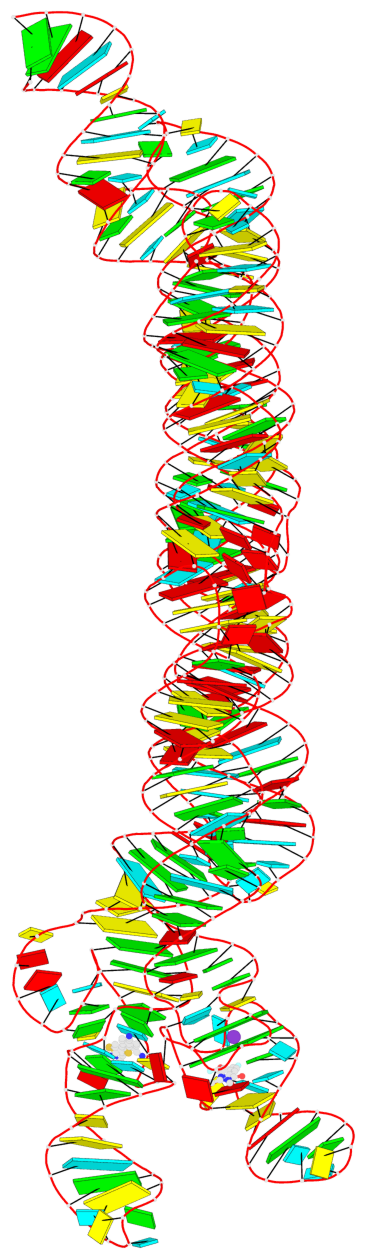
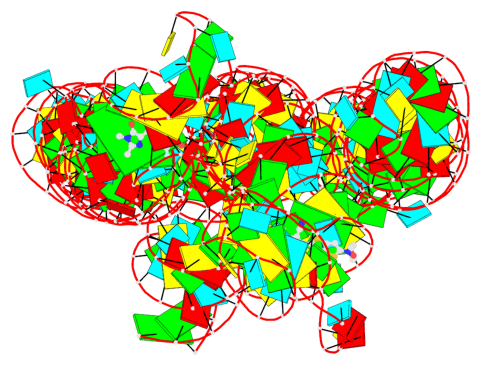
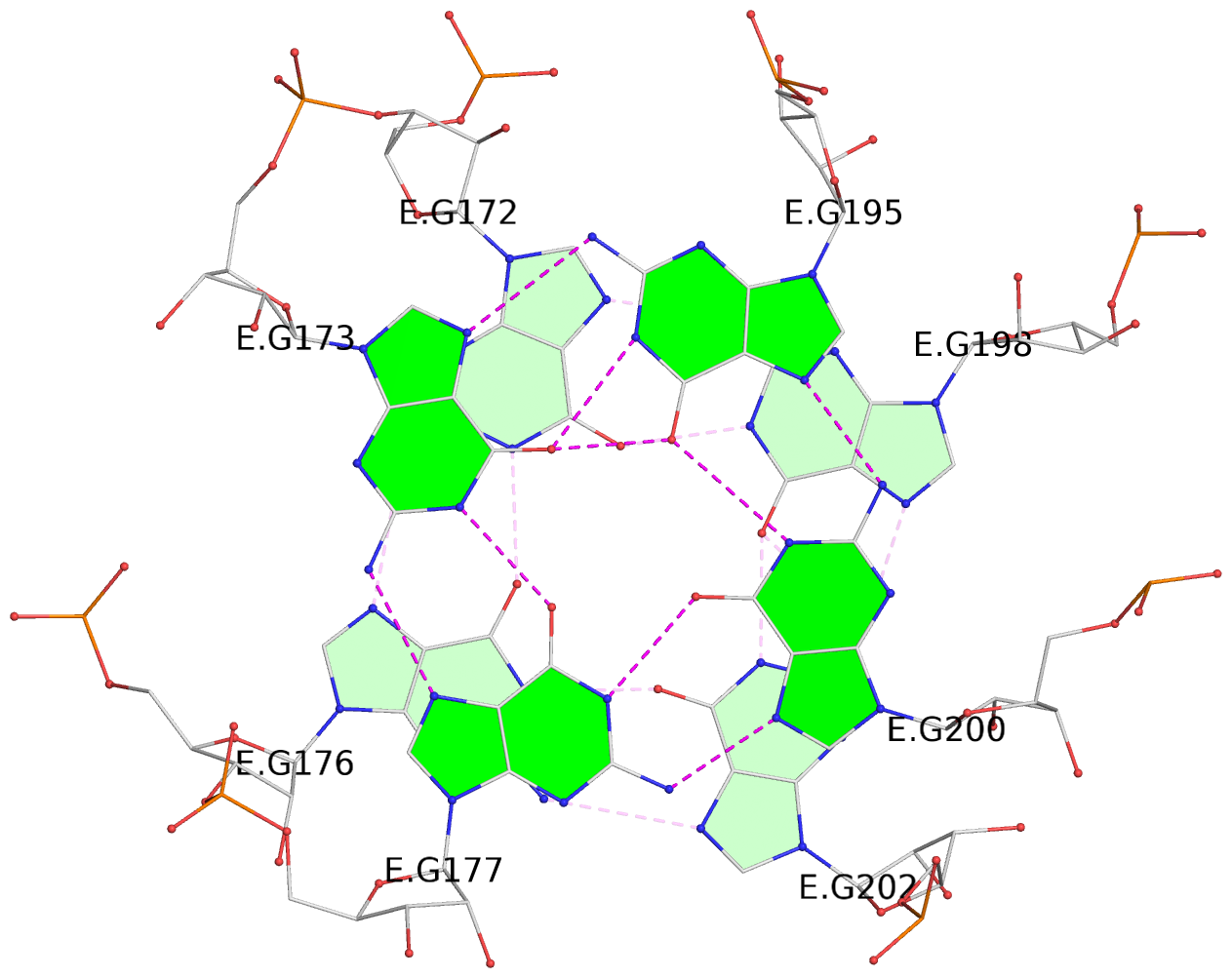
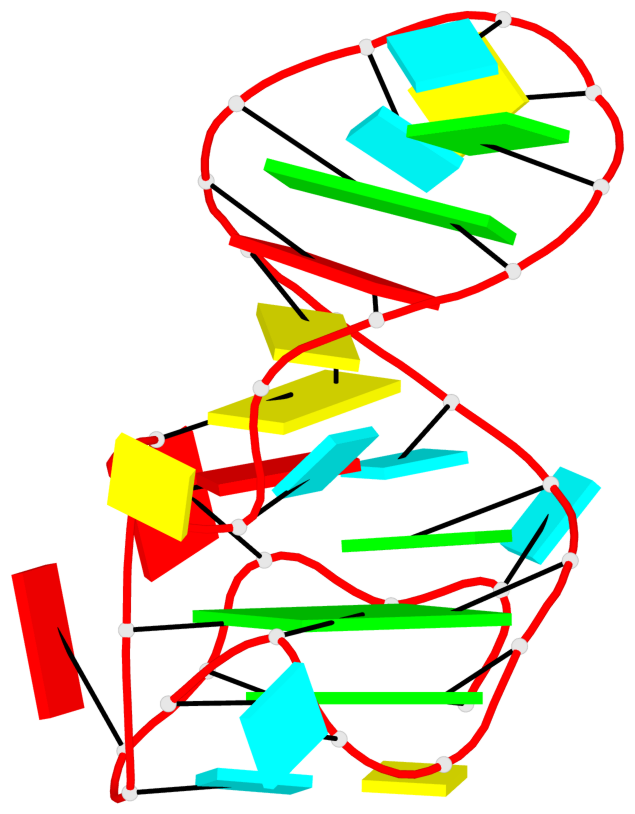
Detailed DSSR results for the G-quadruplex: PDB entry 7zj4
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7zj4
- Class
- RNA
- Method
- cryo-EM (4.43 Å)
- Summary
- Ligand bound state of a brocolli-pepper aptamer fret tile
- Reference
- McRae EKS, Vallina NS, Hansen BK, Boussebayle A, Andersen ES: "Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding."
- Abstract
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-PD+P), (2+2), UUDD
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=--s- sugar=-333 groove=-wn- planarity=0.297 type=other nts=4 GGGG E.G172,E.G176,E.G202,E.G198 2 glyco-bond=--ss sugar=-3-3 groove=-w-n planarity=0.230 type=other nts=4 GGGG E.G173,E.G177,E.G200,E.G195
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.