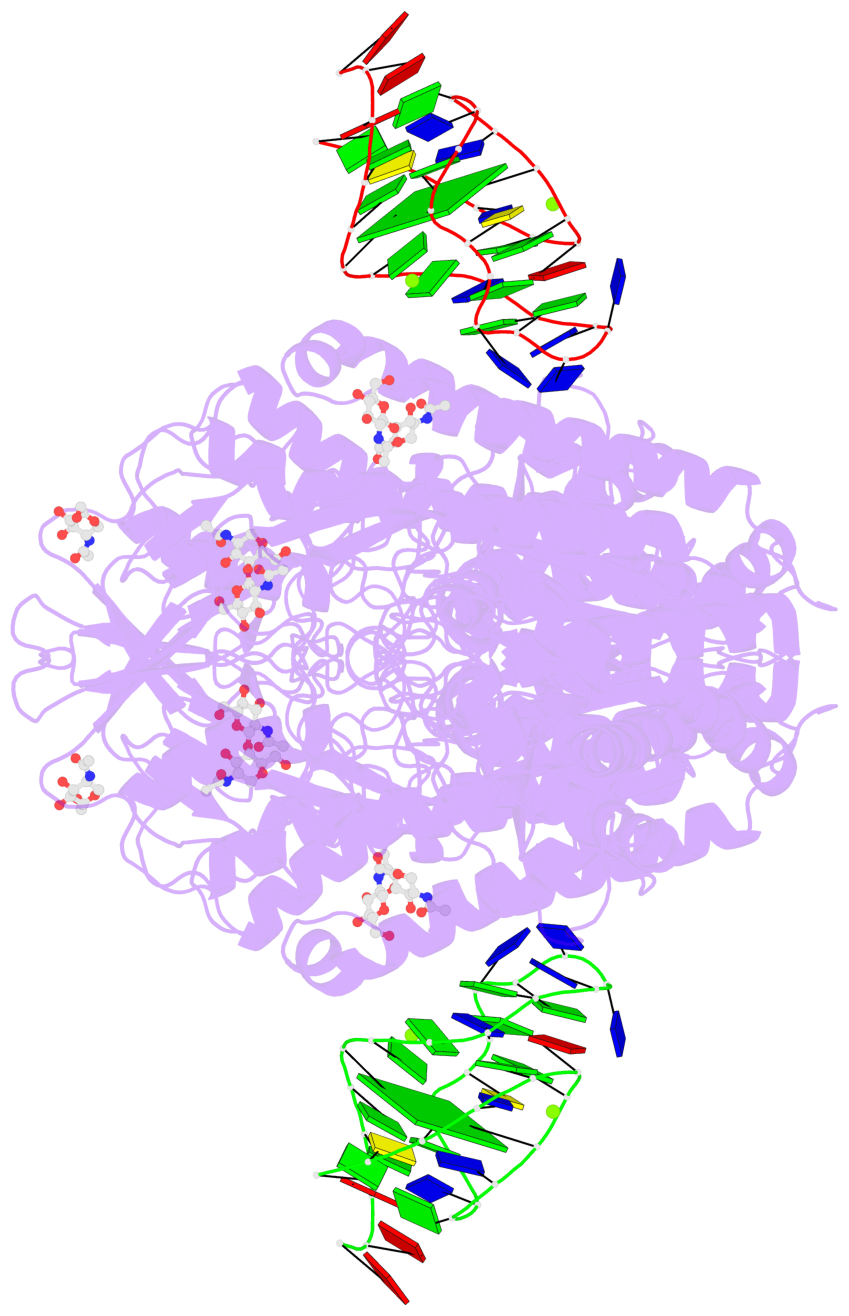
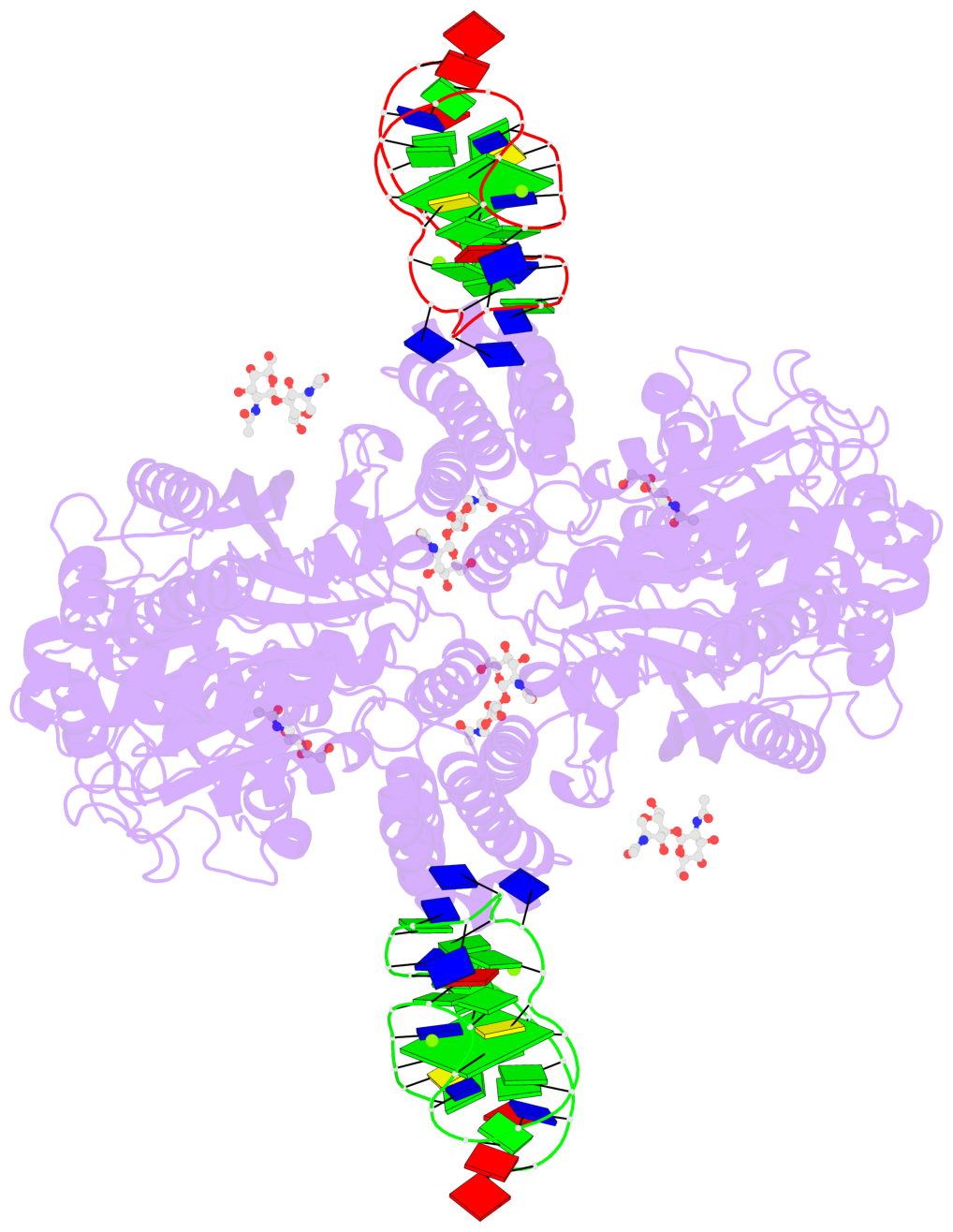
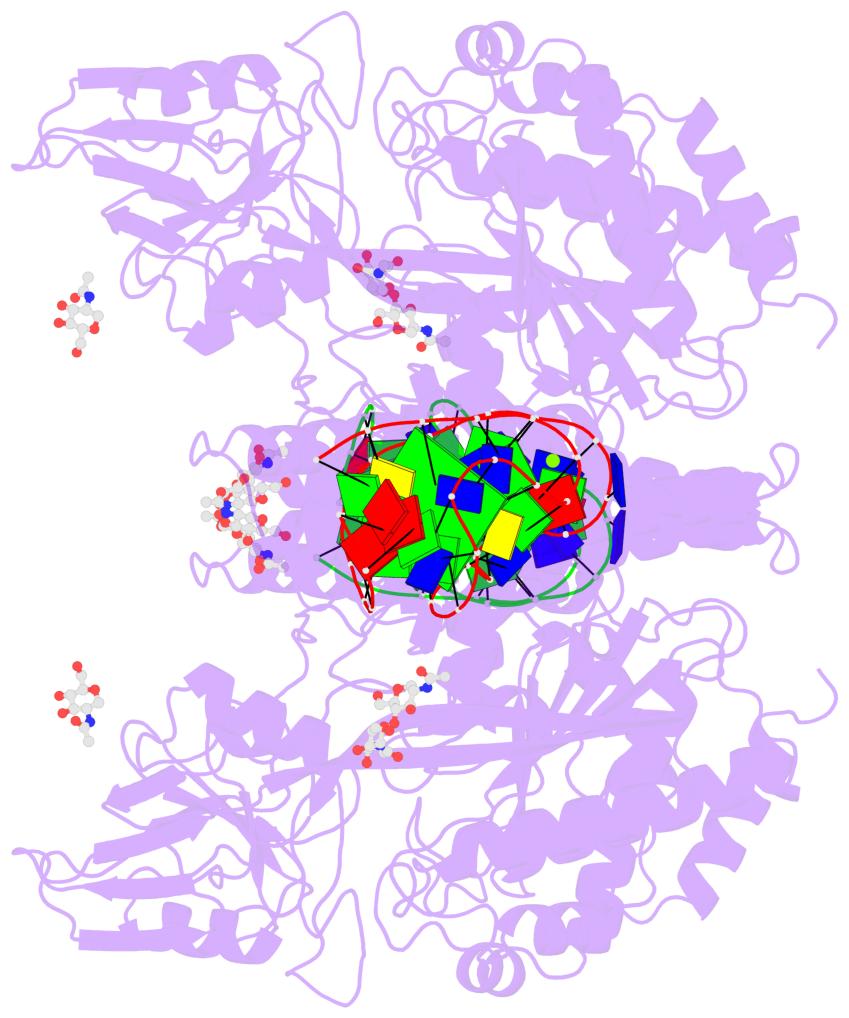
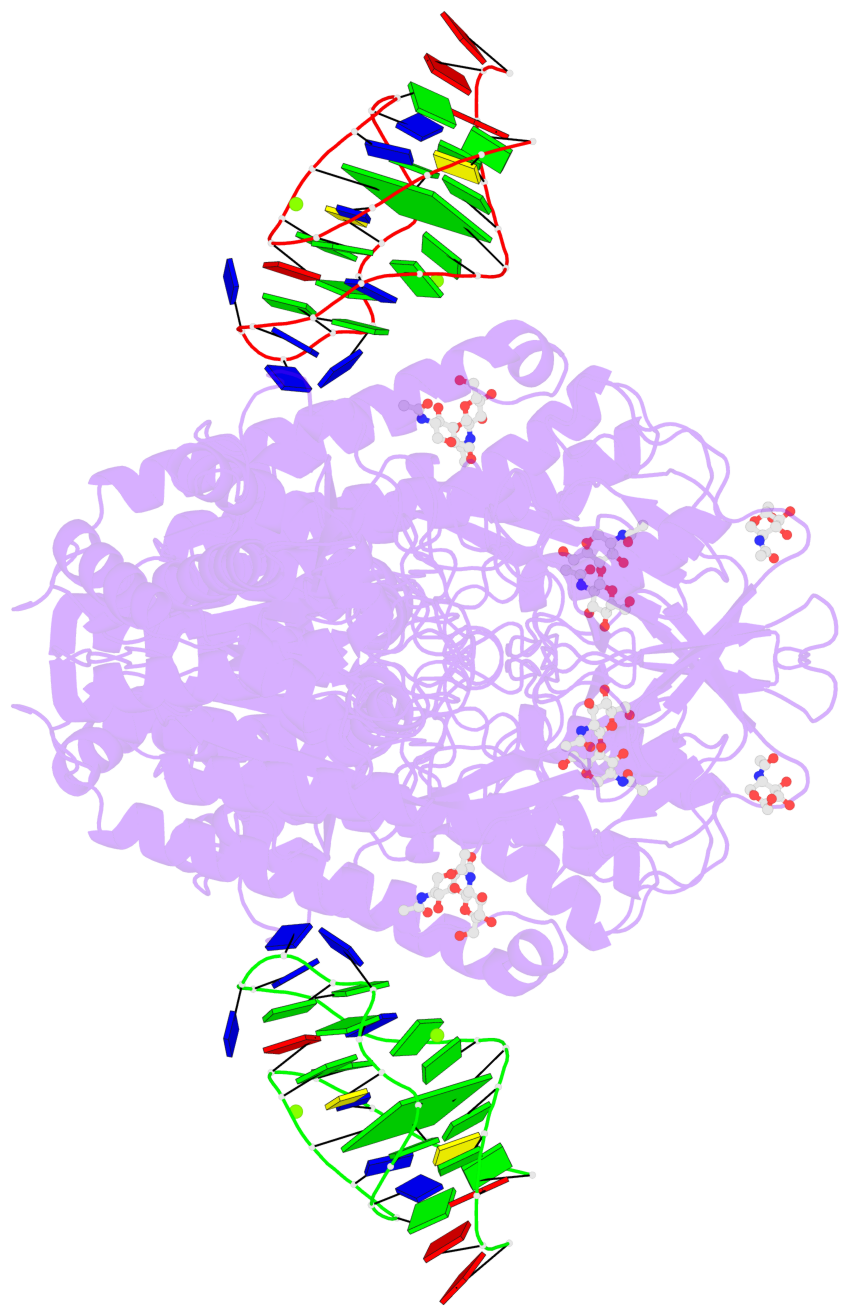
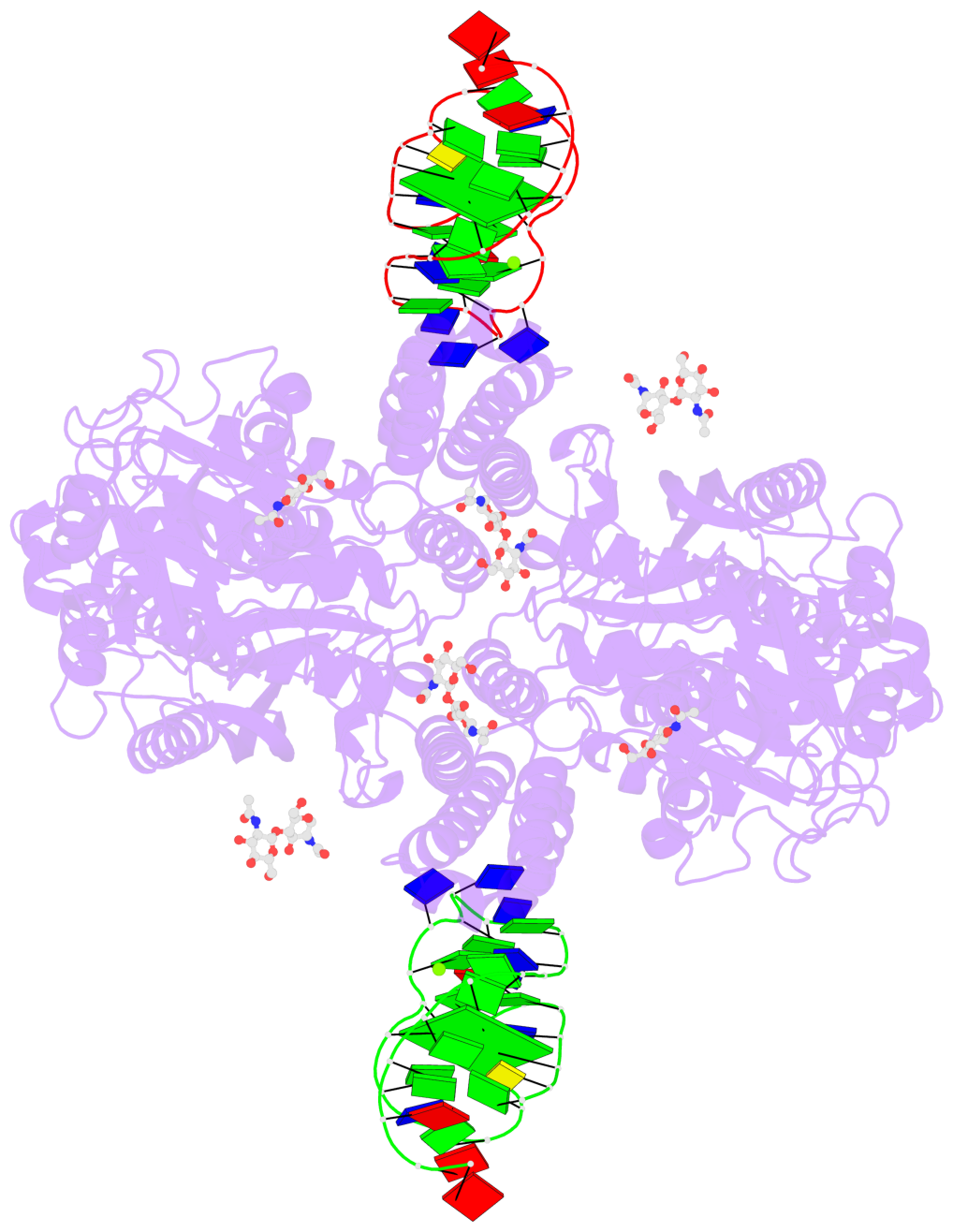
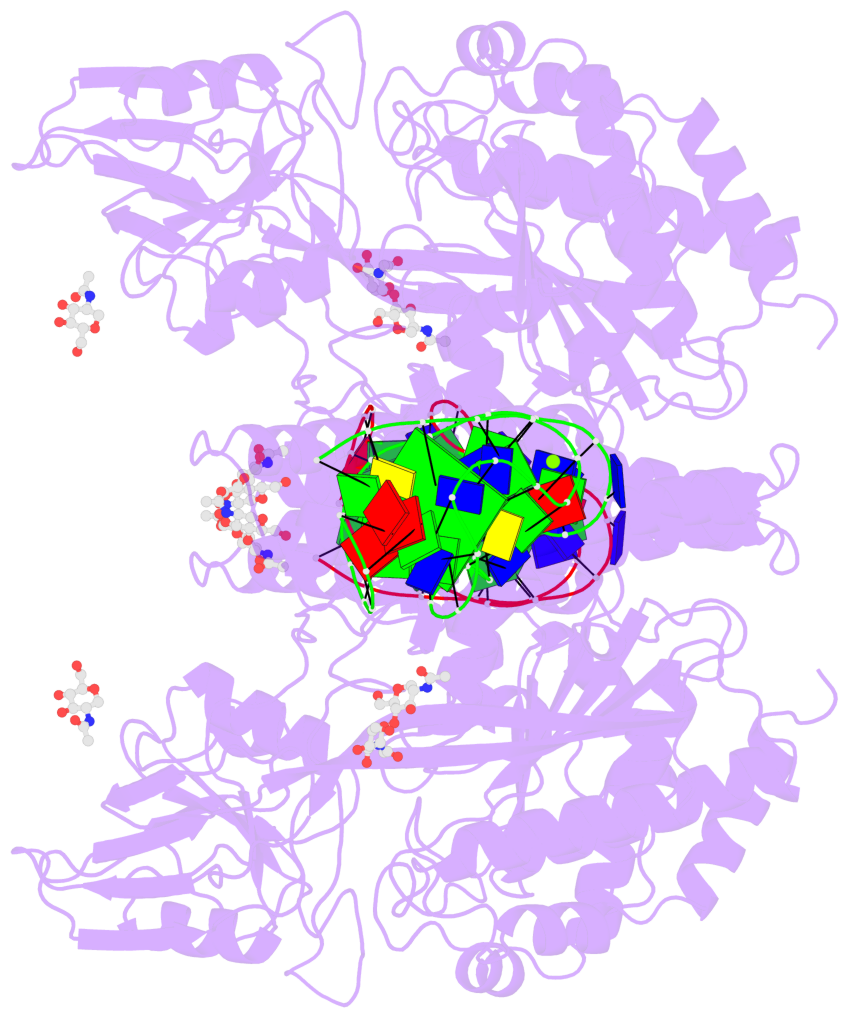
Detailed DSSR results for the G-quadruplex: PDB entry 7zqs
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7zqs
- Class
- membrane protein
- Method
- cryo-EM (2.54 Å)
- Summary
- cryo-EM structure of human transferrin receptor 1 bound to DNA aptamer
- Reference
- Cheng EL, Cardle II, Kacherovsky N, Bansia H, Wang T, Zhou Y, Raman J, Yen A, Gutierrez D, Salipante SJ, des Georges A, Jensen MC, Pun SH (2022): "Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing." J.Am.Chem.Soc., 144, 13851-13864. doi: 10.1021/jacs.2c05349.
- Abstract
- The clinical manufacturing of chimeric antigen receptor (CAR) T cells includes cell selection, activation, gene transduction, and expansion. While the method of T-cell selection varies across companies, current methods do not actively eliminate the cancer cells in the patient's apheresis product from the healthy immune cells. Alarmingly, it has been found that transduction of a single leukemic B cell with the CAR gene can confer resistance to CAR T-cell therapy and lead to treatment failure. In this study, we report the identification of a novel high-affinity DNA aptamer, termed tJBA8.1, that binds transferrin receptor 1 (TfR1), a receptor broadly upregulated by cancer cells. Using competition assays, high resolution cryo-EM, and de novo model building of the aptamer into the resulting electron density, we reveal that tJBA8.1 shares a binding site on TfR1 with holo-transferrin, the natural ligand of TfR1. We use tJBA8.1 to effectively deplete B lymphoma cells spiked into peripheral blood mononuclear cells with minimal impact on the healthy immune cell composition. Lastly, we present opportunities for affinity improvement of tJBA8.1. As TfR1 expression is broadly upregulated in many cancers, including difficult-to-treat T-cell leukemias and lymphomas, our work provides a facile, universal, and inexpensive approach for comprehensively removing cancerous cells from patient apheresis products for safe manufacturing of adoptive T-cell therapies.
- G4 notes
- 2 G-tetrads
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.624 type=saddle nts=4 GGGG A.DG16,A.DG38,A.DG33,A.DG28 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.624 type=saddle nts=4 GGGG C.DG16,C.DG38,C.DG33,C.DG28
List of 6 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#-1 nts=13 GGGTGTTTGTGCG A.DG16,A.DG17,A.DG18,A.DT19,A.DG20,A.DT21,A.DT22,A.DT23,A.DG24,A.DT25,A.DG26,A.DC27,A.DG28 2 type=lateral helix=#-1 nts=6 GGTGTG A.DG28,A.DG29,A.DT30,A.DG31,A.DT32,A.DG33 3 type=lateral helix=#-1 nts=6 GGAGTG A.DG33,A.DG34,A.DA35,A.DG36,A.DT37,A.DG38 4 type=lateral helix=#-2 nts=13 GGGTGTTTGTGCG C.DG16,C.DG17,C.DG18,C.DT19,C.DG20,C.DT21,C.DT22,C.DT23,C.DG24,C.DT25,C.DG26,C.DC27,C.DG28 5 type=lateral helix=#-2 nts=6 GGTGTG C.DG28,C.DG29,C.DT30,C.DG31,C.DT32,C.DG33 6 type=lateral helix=#-2 nts=6 GGAGTG C.DG33,C.DG34,C.DA35,C.DG36,C.DT37,C.DG38