Detailed DSSR results for the G-quadruplex: PDB entry 7zur
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7zur
- Class
- DNA
- Method
- X-ray (1.6 Å)
- Summary
- Four carbons pendant pyridine derivative of the natural alkaloid berberine as human telomeric g-quadruplex binder
- Reference
- Bazzicalupi C, Bonardi A, Biver T, Ferraroni M, Papi F, Savastano M, Lombardi P, Gratteri P (2022): "Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis." Int J Mol Sci, 23. doi: 10.3390/ijms232214061.
- Abstract
- The interaction between the series of berberine derivatives
- G4 notes
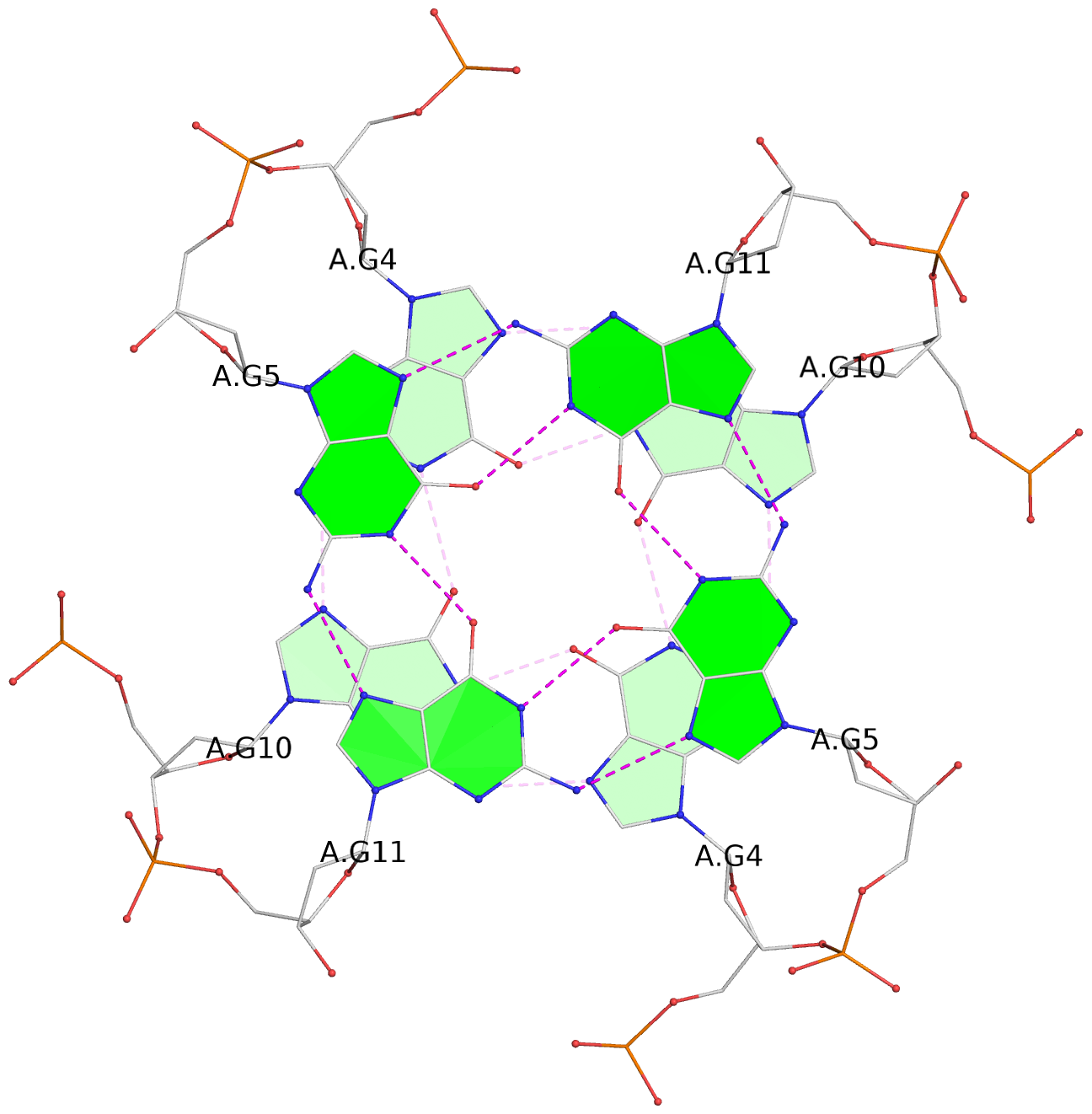
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
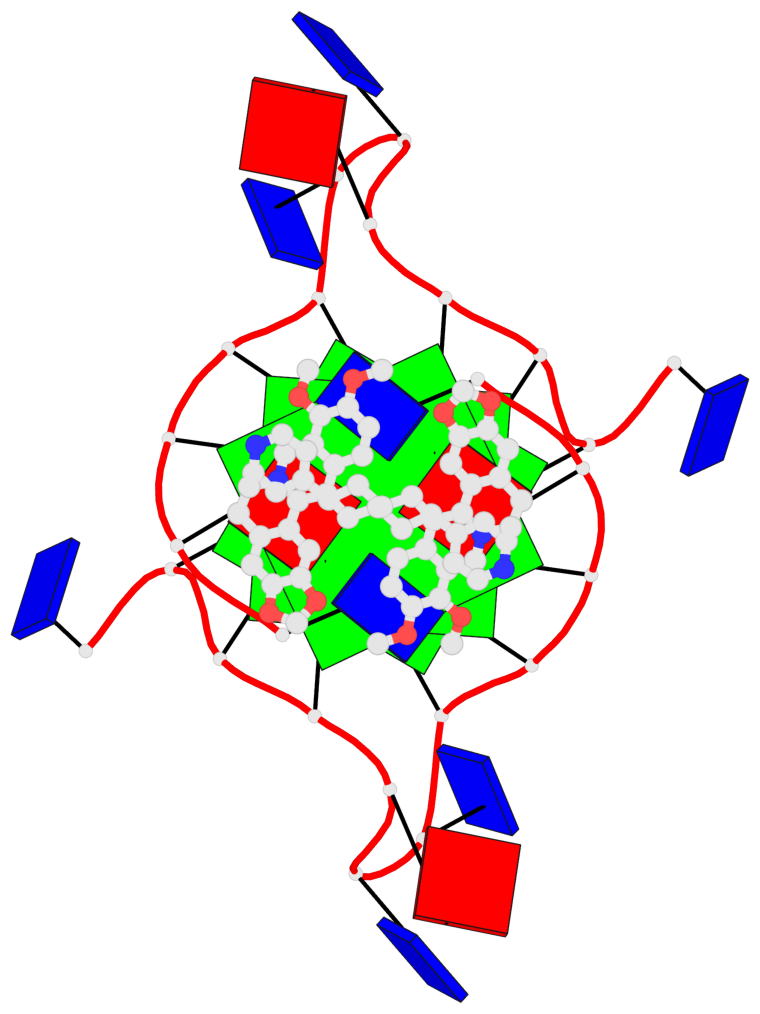
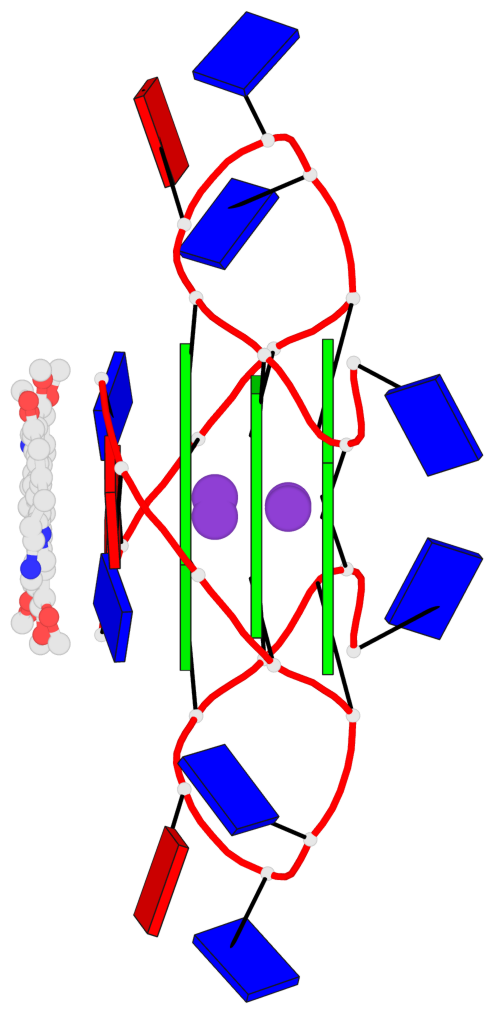
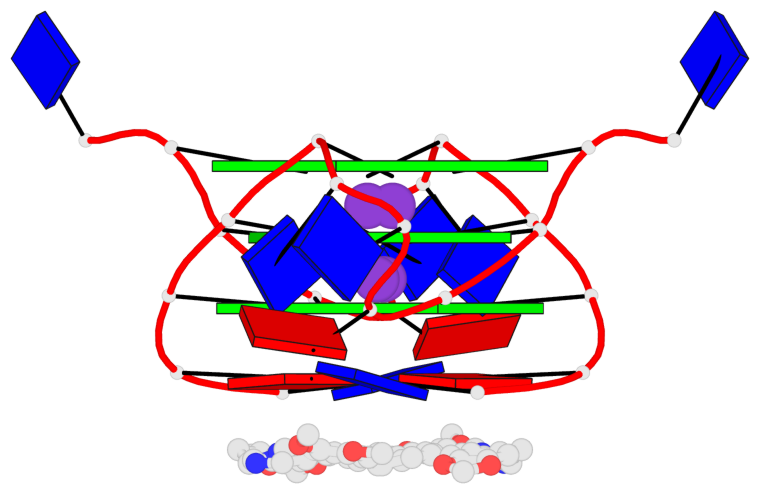
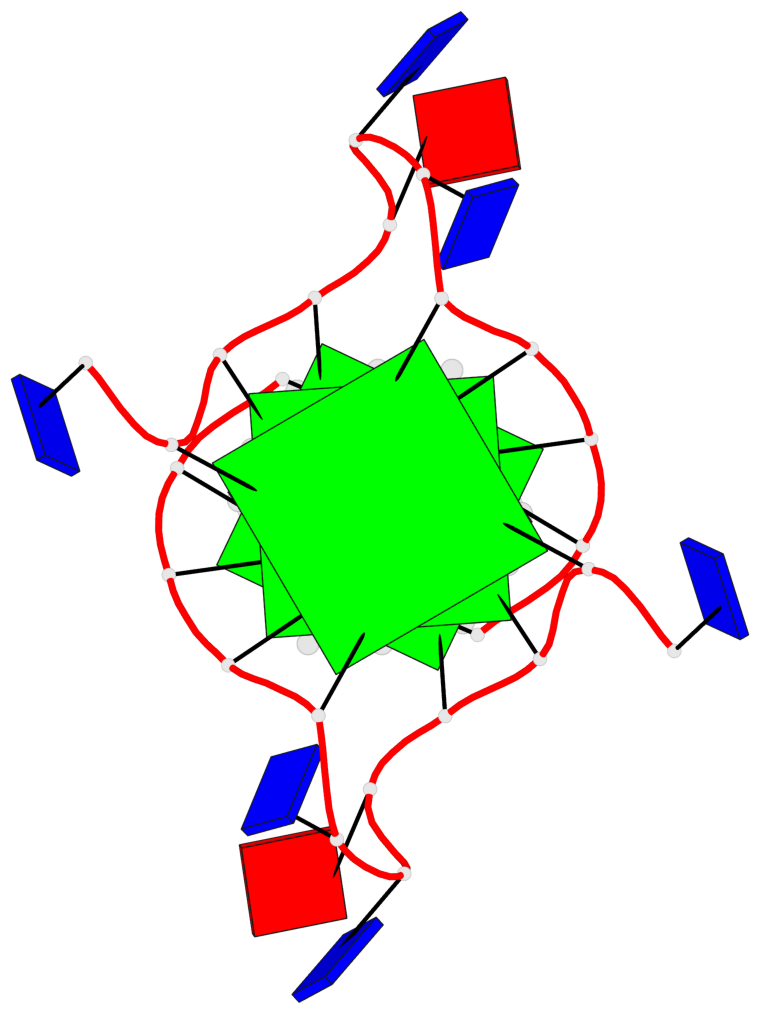
Base-block schematics in six views
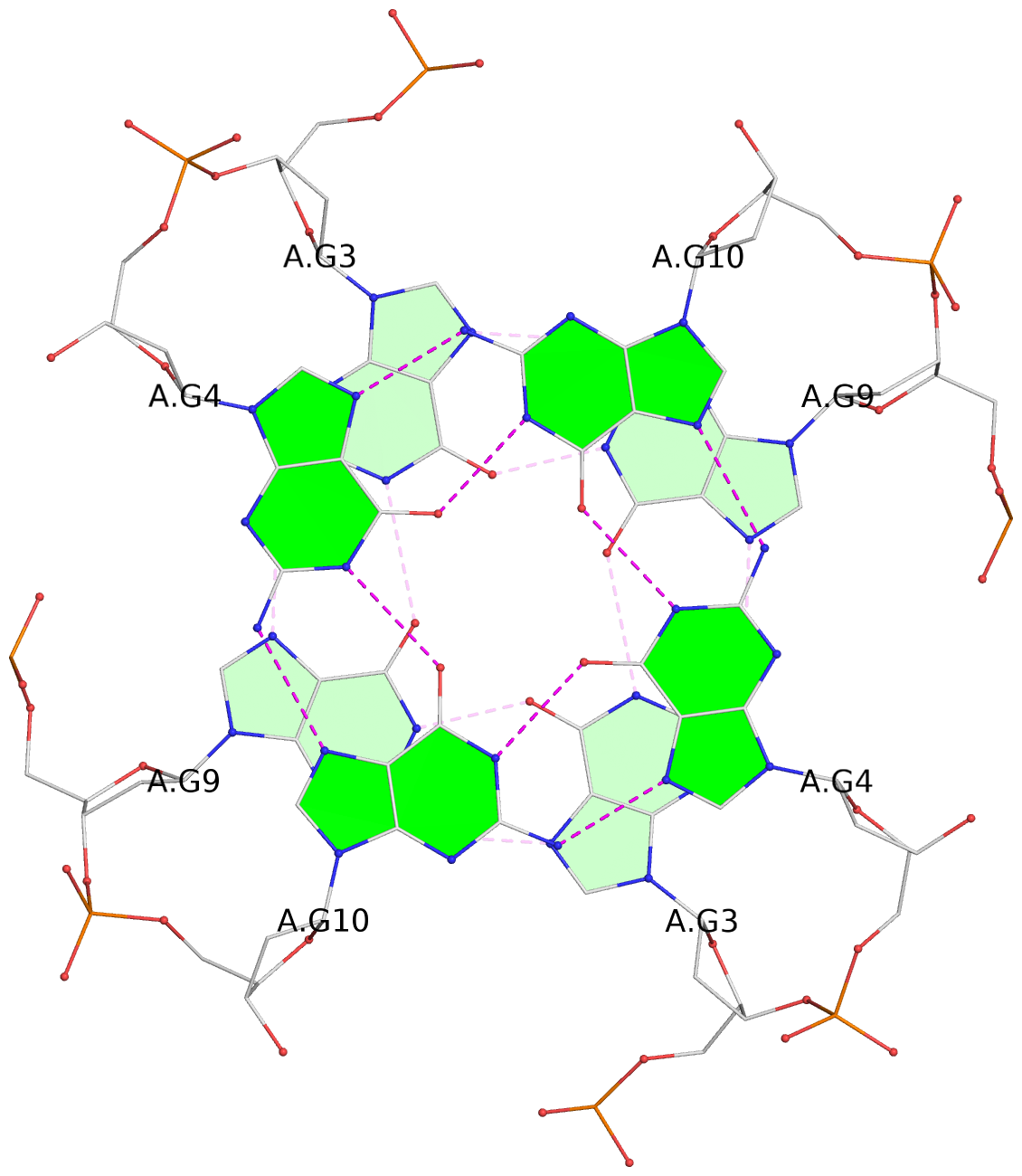
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.128 type=planar nts=4 GGGG 1:A.DG3,1:A.DG9,2:A.DG3,2:A.DG9 2 glyco-bond=---- sugar=---- groove=---- planarity=0.149 type=planar nts=4 GGGG 1:A.DG4,1:A.DG10,2:A.DG4,2:A.DG10 3 glyco-bond=---- sugar=---- groove=---- planarity=0.300 type=bowl nts=4 GGGG 1:A.DG5,1:A.DG11,2:A.DG5,2:A.DG11
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.