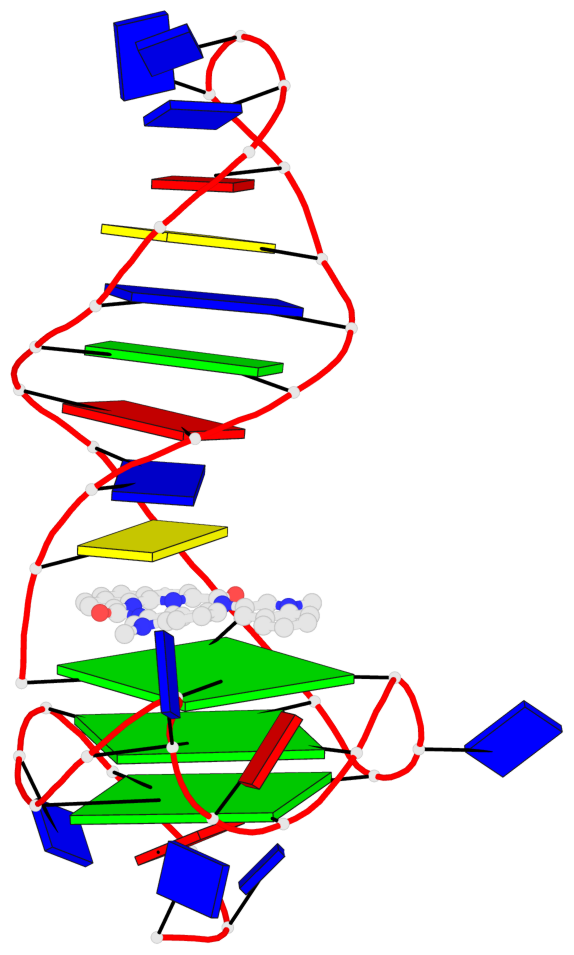
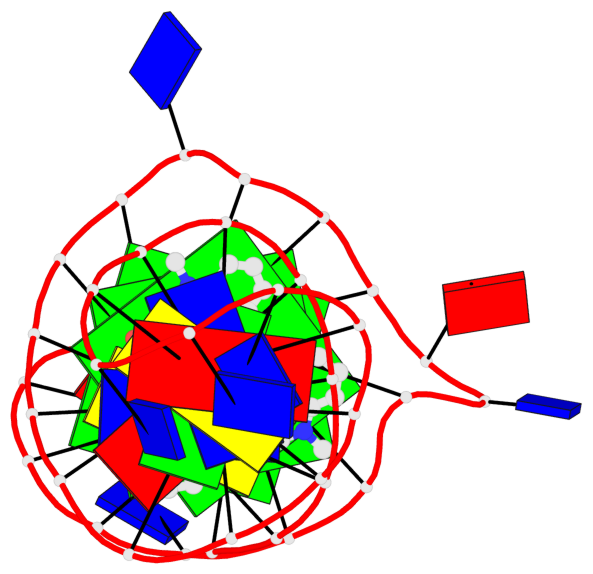
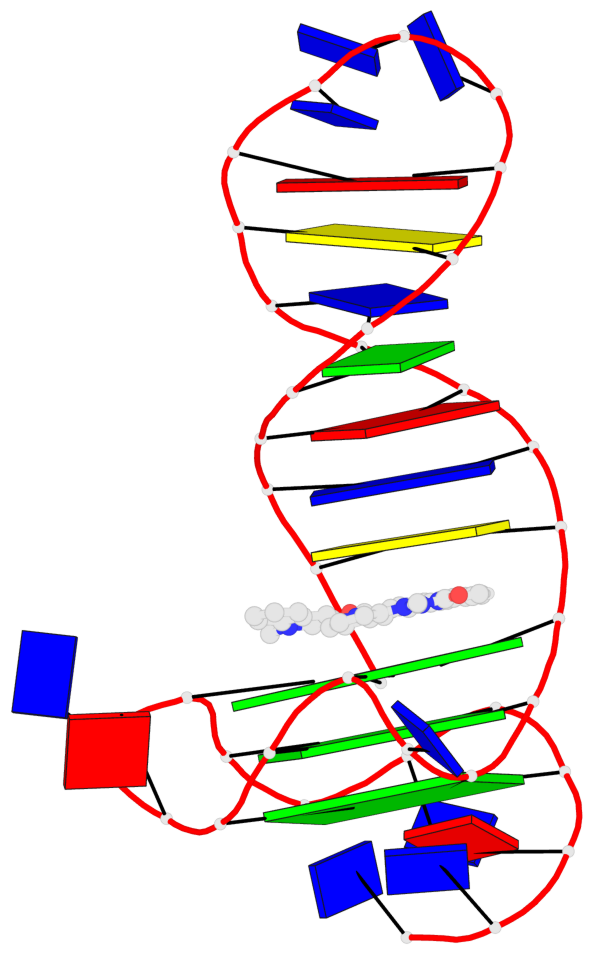
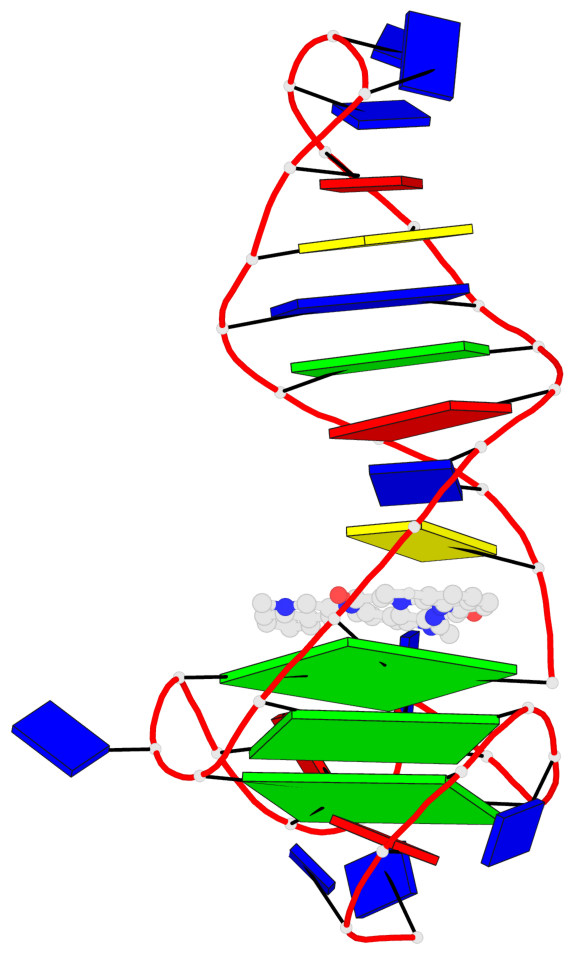
Detailed DSSR results for the G-quadruplex: PDB entry 8abd
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8abd
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of phen-dc3 intercalating into a quadruplex-duplex hybrid
- Reference
- Vianney YM, Weisz K (2022): "High-affinity binding at quadruplex-duplex junctions: rather the rule than the exception." Nucleic Acids Res., 50, 11948-11964. doi: 10.1093/nar/gkac1088.
- Abstract
- Quadruplex-duplex (Q-D) junctions constitute unique structural motifs in genomic sequences. Through comprehensive calorimetric as well as high-resolution NMR structural studies, Q-D junctions with a hairpin-type snapback loop coaxially stacked onto an outer G-tetrad were identified to be most effective binding sites for various polycyclic quadruplex ligands. The Q-D interface is readily recognized by intercalation of the ligand aromatic core structure between G-tetrad and the neighboring base pair. Based on the thermodynamic and structural data, guidelines for the design of ligands with enhanced selectivity towards a Q-D interface emerge. Whereas intercalation at Q-D junctions mostly outcompete stacking at the quadruplex free outer tetrad or intercalation between duplex base pairs to varying degrees, ligand side chains considerably contribute to the selectivity for a Q-D target over other binding sites. In contrast to common perceptions, an appended side chain that additionally interacts within the duplex minor groove may confer only poor selectivity. Rather, the Q-D selectivity is suggested to benefit from an extension of the side chain towards the exposed part of the G-tetrad at the junction. The presented results will support the design of selective high-affinity binding ligands for targeting Q-D interfaces in medicinal but also technological applications.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
Base-block schematics in six views
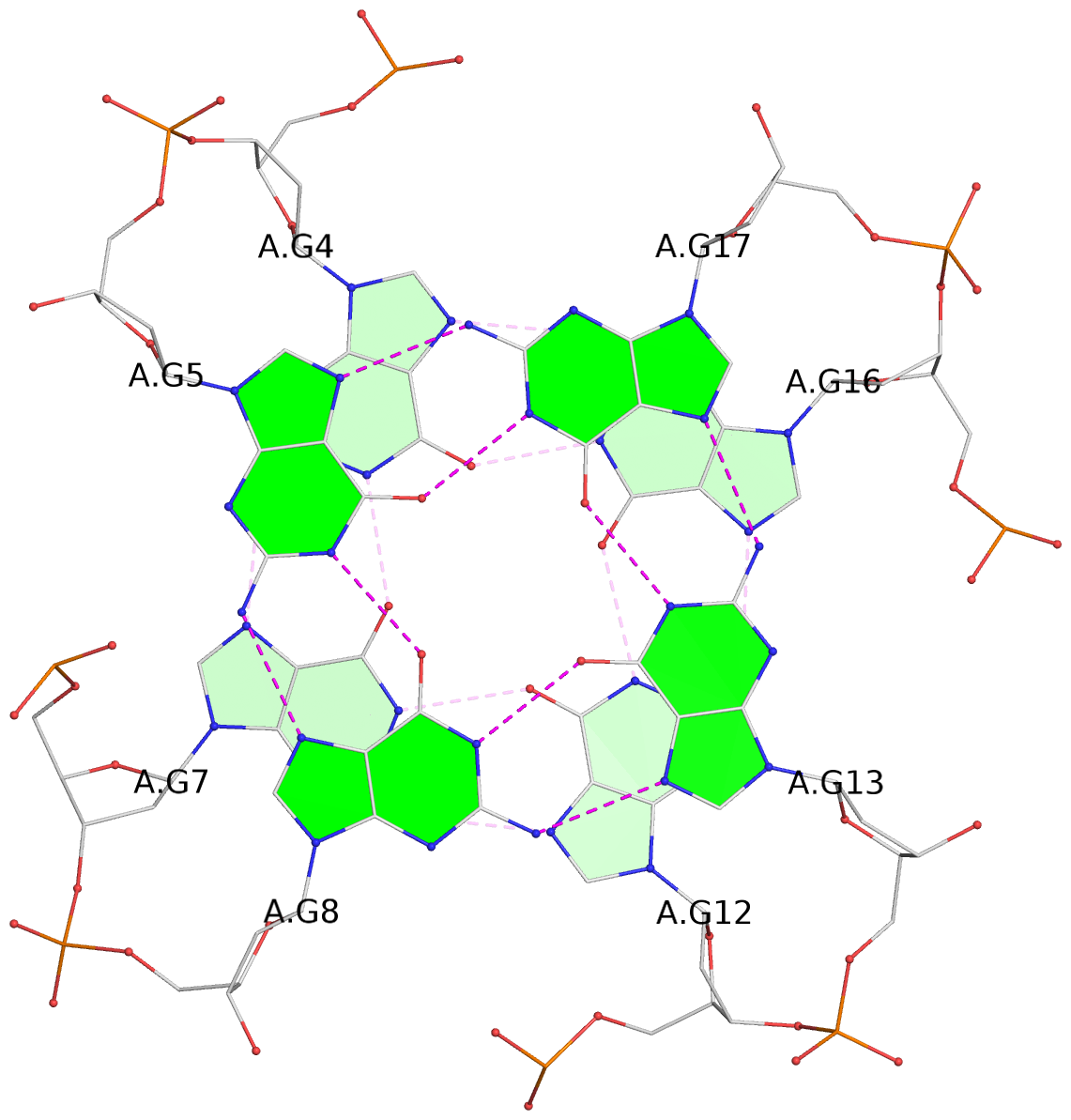
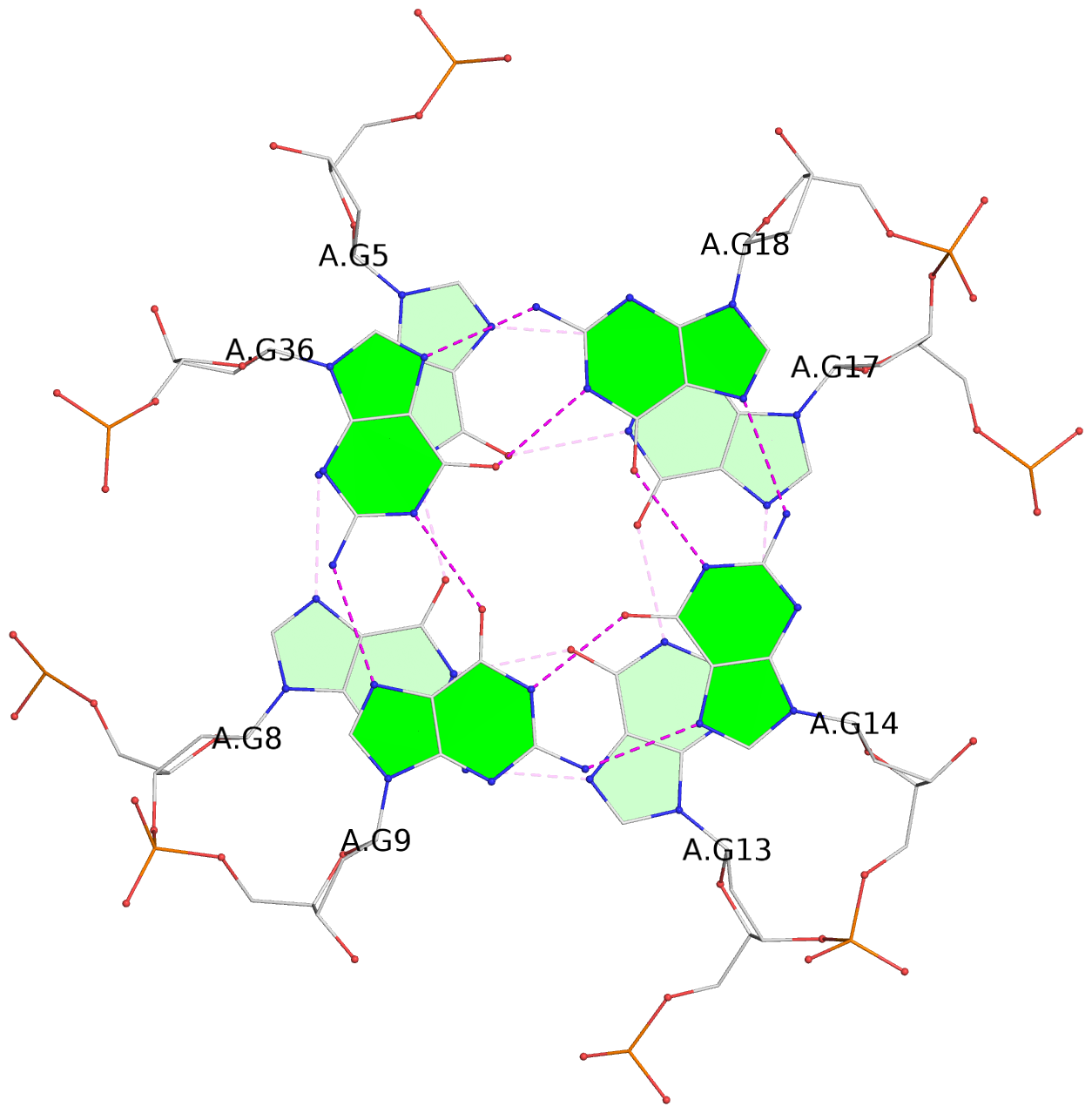
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.257 type=other nts=4 GGGG A.DG4,A.DG7,A.DG12,A.DG16 2 glyco-bond=---- sugar=---- groove=---- planarity=0.144 type=planar nts=4 GGGG A.DG5,A.DG8,A.DG13,A.DG17 3 glyco-bond=---s sugar=---- groove=--wn planarity=0.185 type=other nts=4 GGGG A.DG9,A.DG14,A.DG18,A.DG36
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 non-stem G4-loop (including the two closing Gs)
1 type=lateral helix=#1 nts=19 GCTAGTCATTTTGACTAGG A.DG18,A.DC19,A.DT20,A.DA21,A.DG22,A.DT23,A.DC24,A.DA25,A.DT26,A.DT27,A.DT28,A.DT29,A.DG30,A.DA31,A.DC32,A.DT33,A.DA34,A.DG35,A.DG36