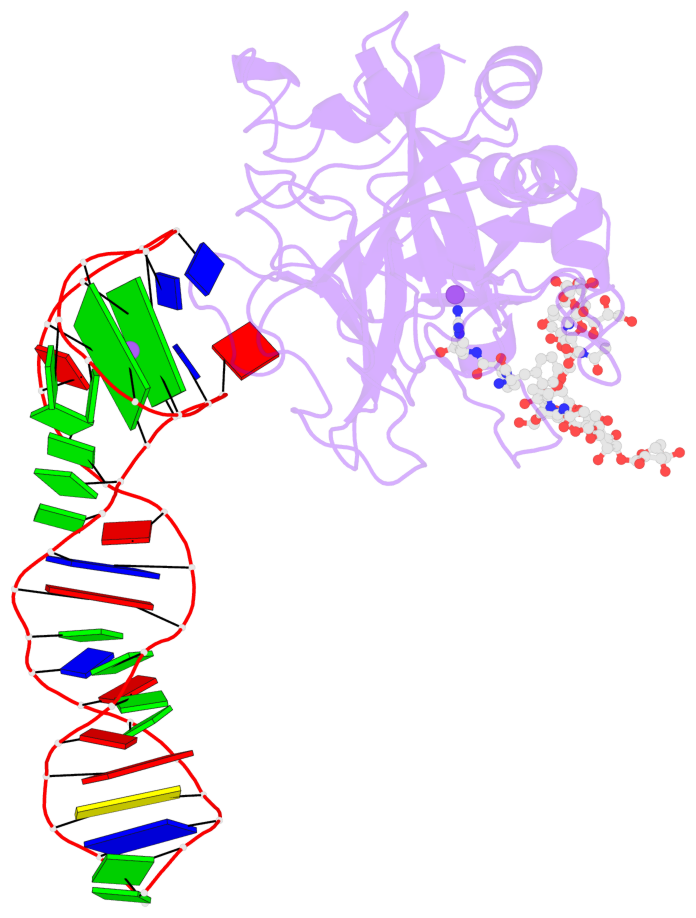
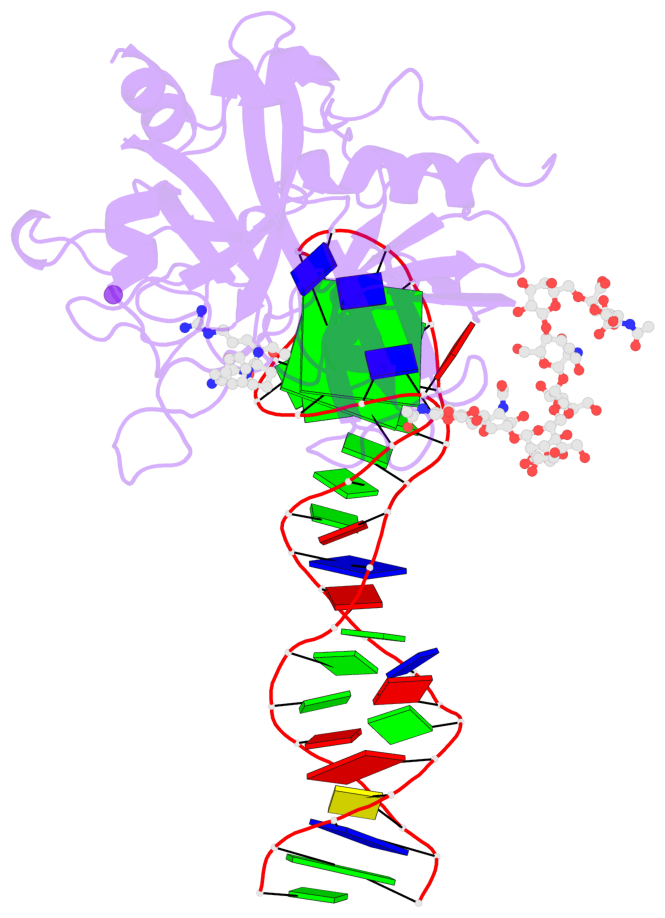
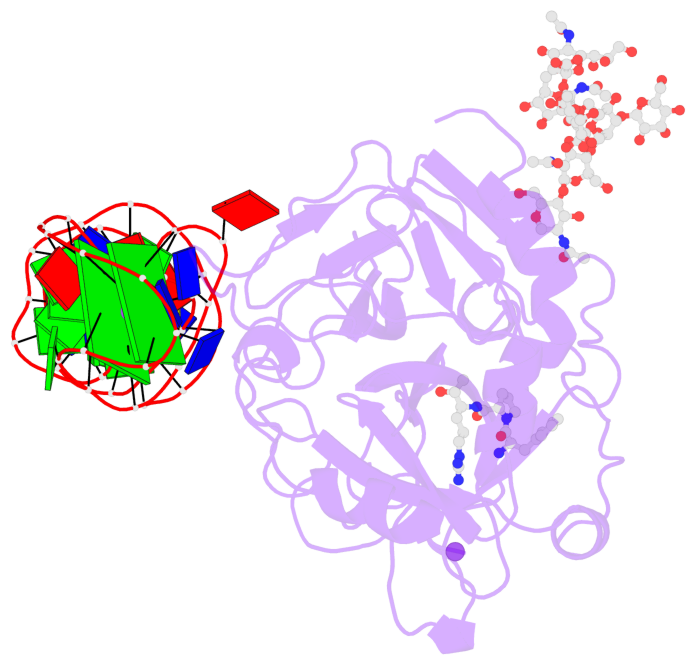
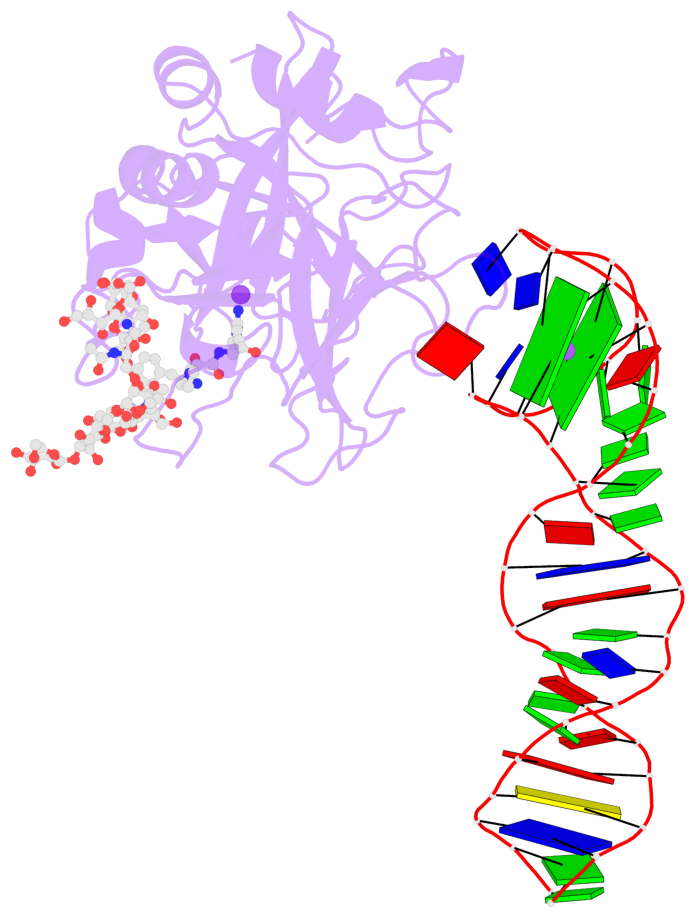
Detailed DSSR results for the G-quadruplex: PDB entry 8bw5
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8bw5
- Class
- hydrolase
- Method
- X-ray (2.8 Å)
- Summary
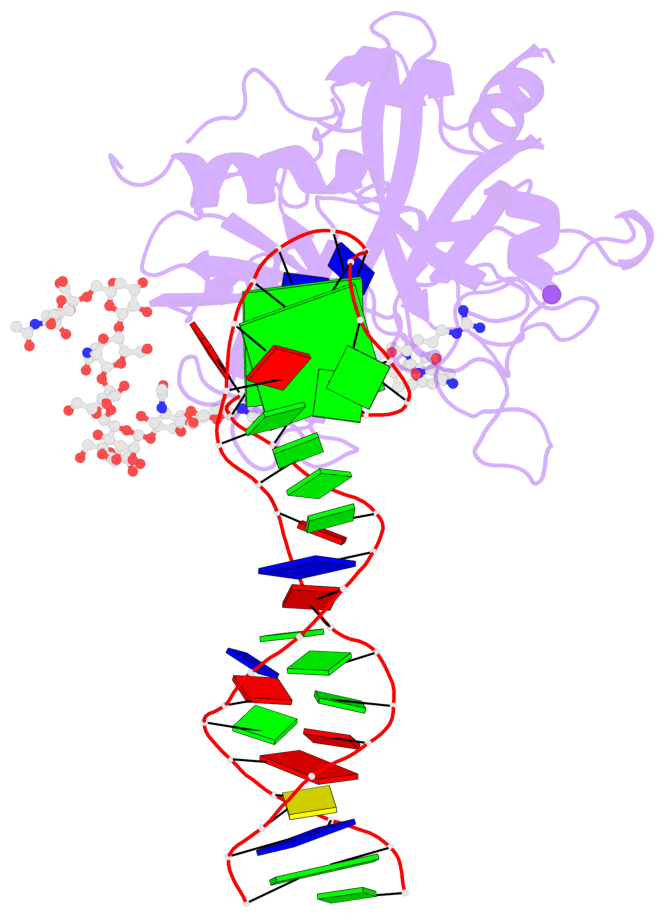
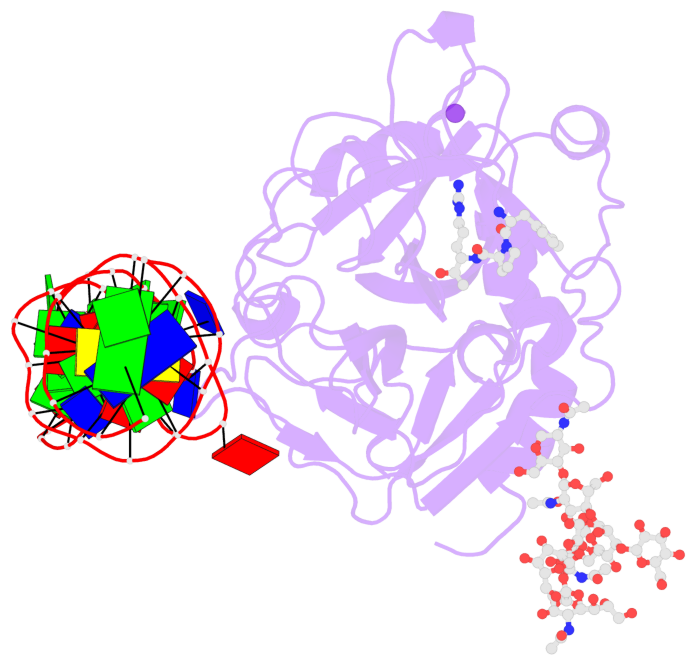
- X-ray structure of the complex between human alpha thrombin and the duplex-quadruplex aptamer m08s-1_41mer
- Reference
- Troisi R, Napolitano V, Rossitto E, Osman W, Nagano M, Wakui K, Popowicz GM, Yoshimoto K, Sica F (2023): "Steric hindrance and structural flexibility shape the functional properties of a guanine-rich oligonucleotide." Nucleic Acids Res., 51, 8880-8890. doi: 10.1093/nar/gkad634.
- Abstract
- Ligand/protein molecular recognition involves a dynamic process, whereby both partners require a degree of structural plasticity to regulate the binding/unbinding event. Here, we present the characterization of the interaction between a highly dynamic G-rich oligonucleotide, M08s-1, and its target protein, human α-thrombin. M08s-1 is the most active anticoagulant aptamer selected thus far. Circular dichroism and gel electrophoresis analyses indicate that both intramolecular and intermolecular G-quadruplex structures are populated in solution. The presence of thrombin stabilises the antiparallel intramolecular chair-like G-quadruplex conformation, that provides by far the main contribution to the biological activity of the aptamer. The crystal structure of the thrombin-oligonucleotide complex reveals that M08s-1 adopts a kinked structural organization formed by a G-quadruplex domain and a long duplex module, linked by a stretch of five purine bases. The quadruplex motif hooks the exosite I region of thrombin and the duplex region is folded towards the surface of the protein. This structural feature, which has never been observed in other anti-exosite I aptamers with a shorter duplex motif, hinders the approach of a protein substrate to the active site region and may well explain the significant increase in the anticoagulant activity of M08s-1 compared to the other anti-exosite I aptamers.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+Ln+Lw+Ln), chair(2+2), UDUD
Base-block schematics in six views
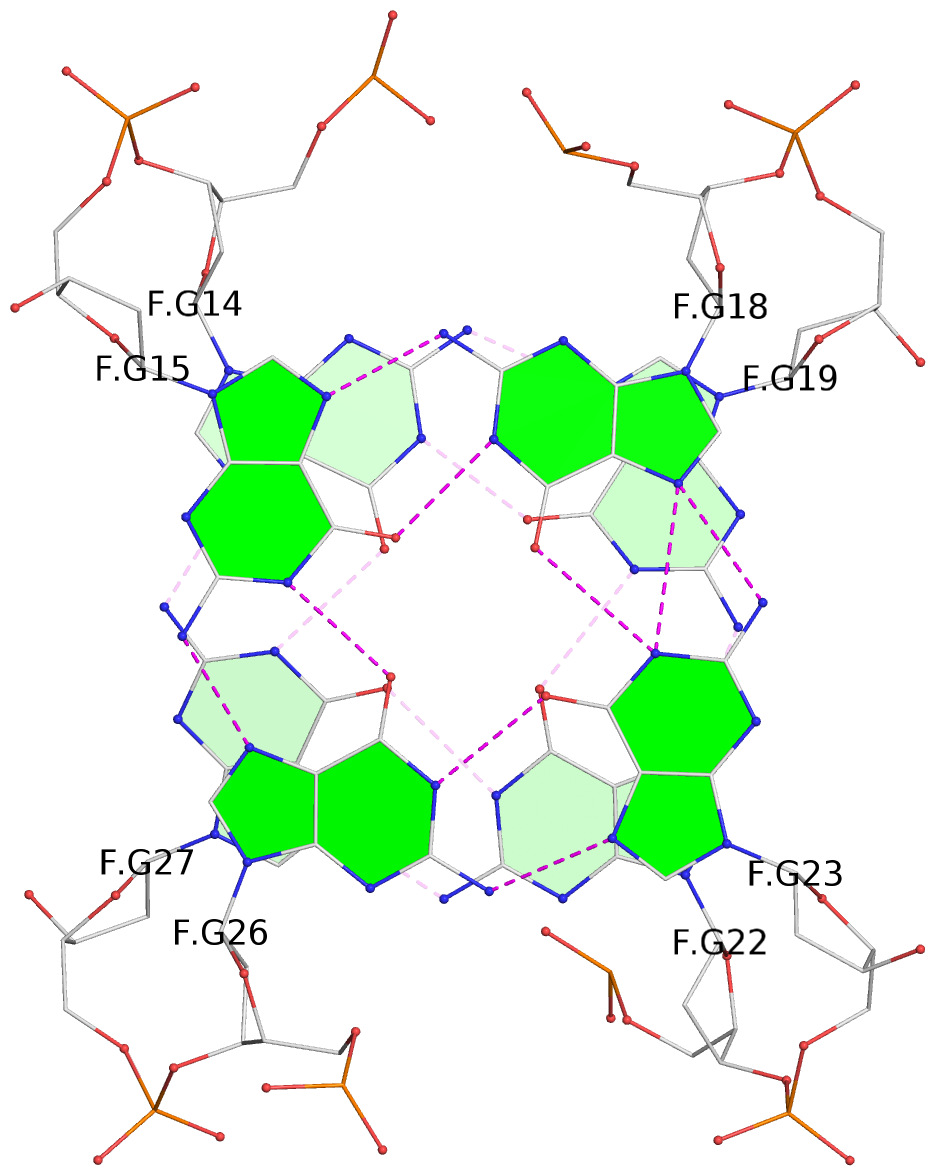
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.550 type=bowl nts=4 GGGG F.DG14,F.DG27,F.DG22,F.DG19 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.537 type=other nts=4 GGGG F.DG15,F.DG26,F.DG23,F.DG18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.