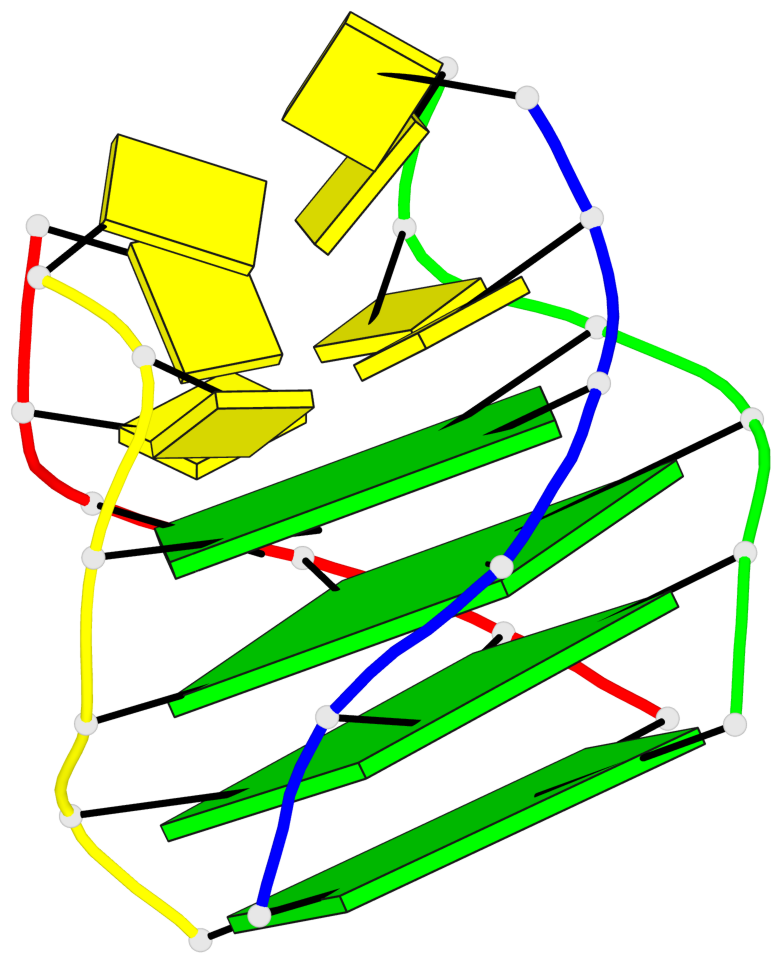
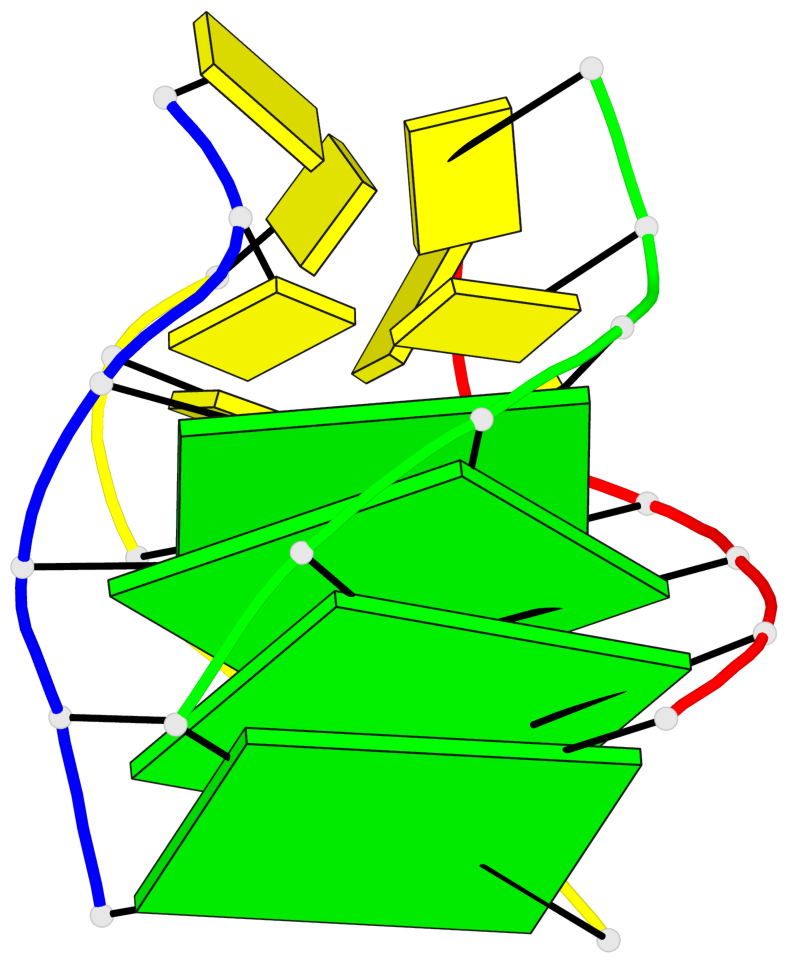
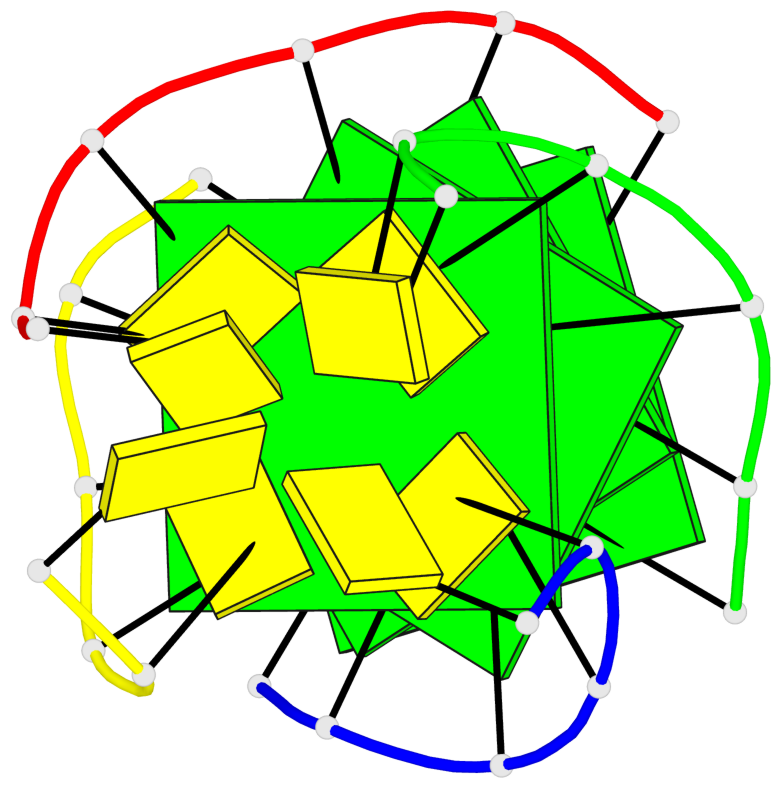
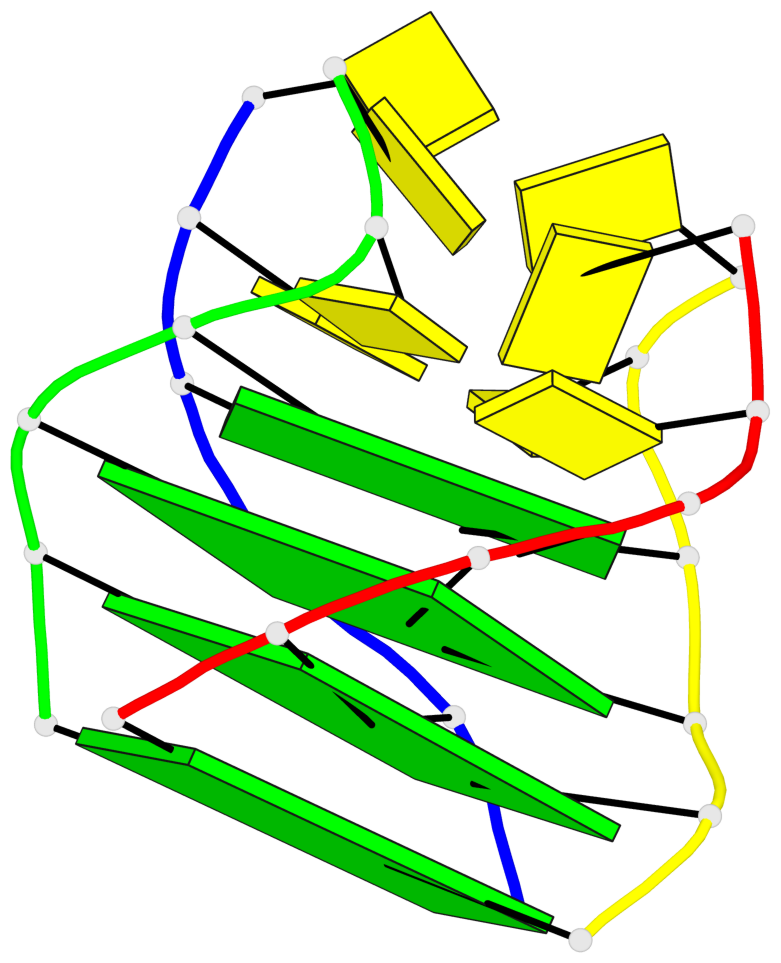
Detailed DSSR results for the G-quadruplex: PDB entry 8c7b
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8c7b
- Class
- DNA
- Method
- NMR
- Summary
- Slow cation movements within tetramolecular g-quadruplex: vacant cation binding sites in addition to all syn g-quartet
- Reference
- Zalar M, Wang B, Plavec J, Sket P (2023): "Insight into Tetramolecular DNA G-Quadruplexes Associated with ALS and FTLD: Cation Interactions and Formation of Higher-Ordered Structure." Int J Mol Sci, 24. doi: 10.3390/ijms241713437.
- Abstract
- The G4C2 hexanucleotide repeat expansion in the c9orf72 gene is a major genetic cause of familial amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), with the formation of G-quadruplexes directly linked to the development of these diseases. Cations play a crucial role in the formation and structure of G-quadruplexes. In this study, we investigated the impact of biologically relevant potassium ions on G-quadruplex structures and utilized 15N-labeled ammonium cations as a substitute for K+ ions to gain further insights into cation binding and exchange dynamics. Through nuclear magnetic resonance spectroscopy and molecular dynamics simulations, we demonstrate that the single d(G4C2) repeat, in the presence of 15NH4+ ions, adopts a tetramolecular G-quadruplex with an all-syn quartet at the 5'-end. The movement of 15NH4+ ions through the central channel of the G-quadruplex, as well as to the bulk solution, is governed by the vacant cation binding site, in addition to the all-syn quartet at the 5'-end. Furthermore, the addition of K+ ions to G-quadruplexes folded in the presence of 15NH4+ ions induces stacking of G-quadruplexes via their 5'-end G-quartets, leading to the formation of stable higher-ordered species.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
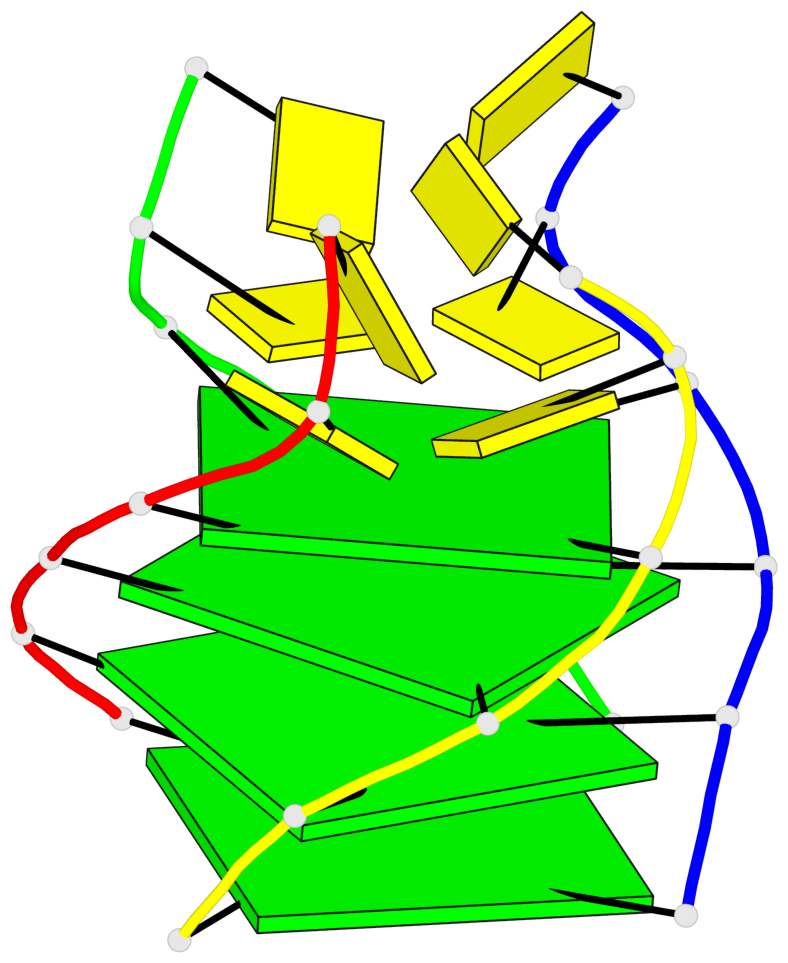
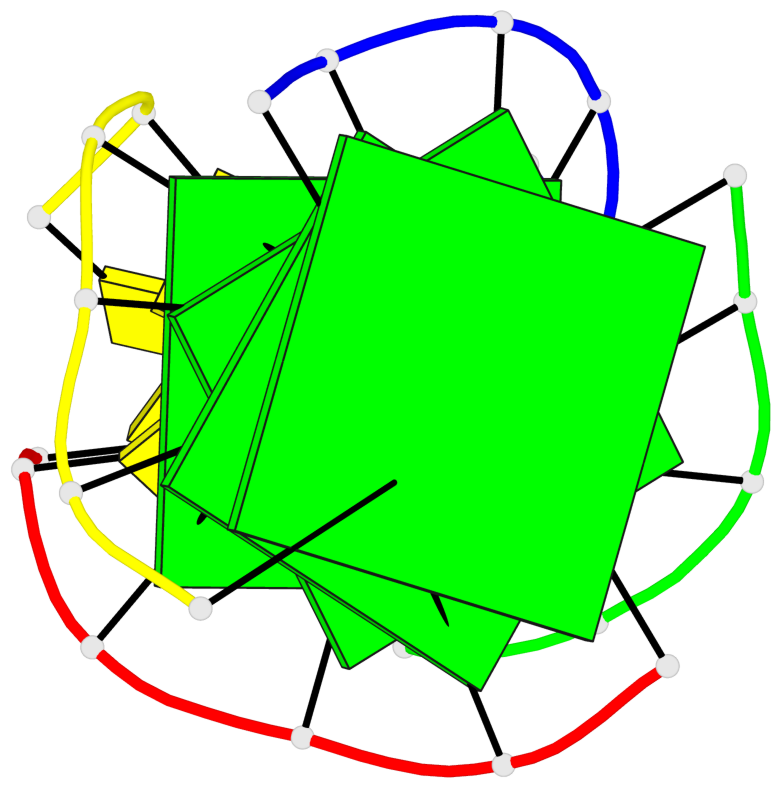
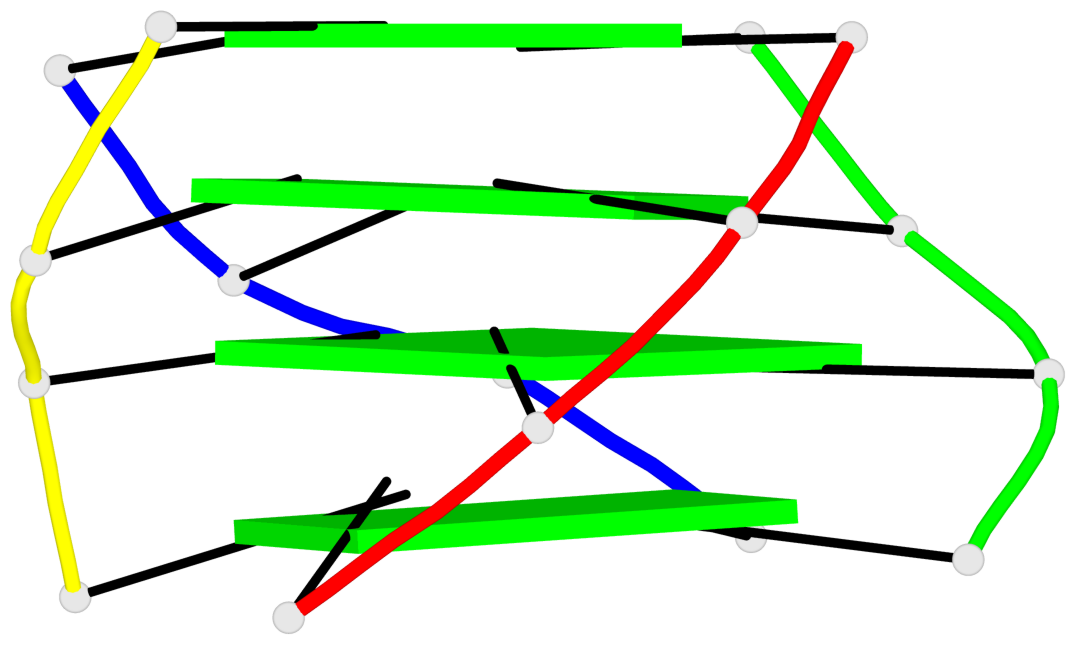
Base-block schematics in six views
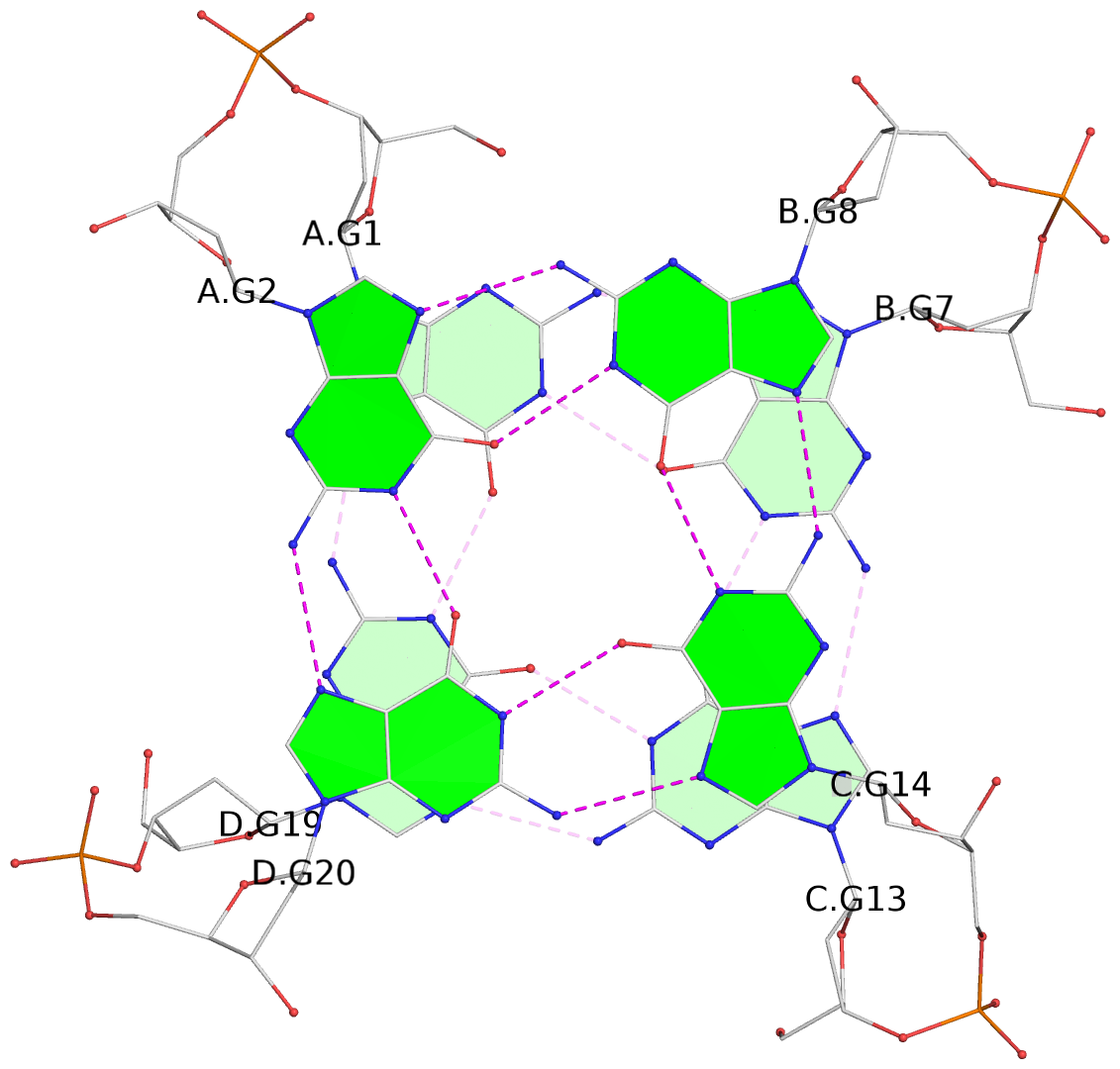
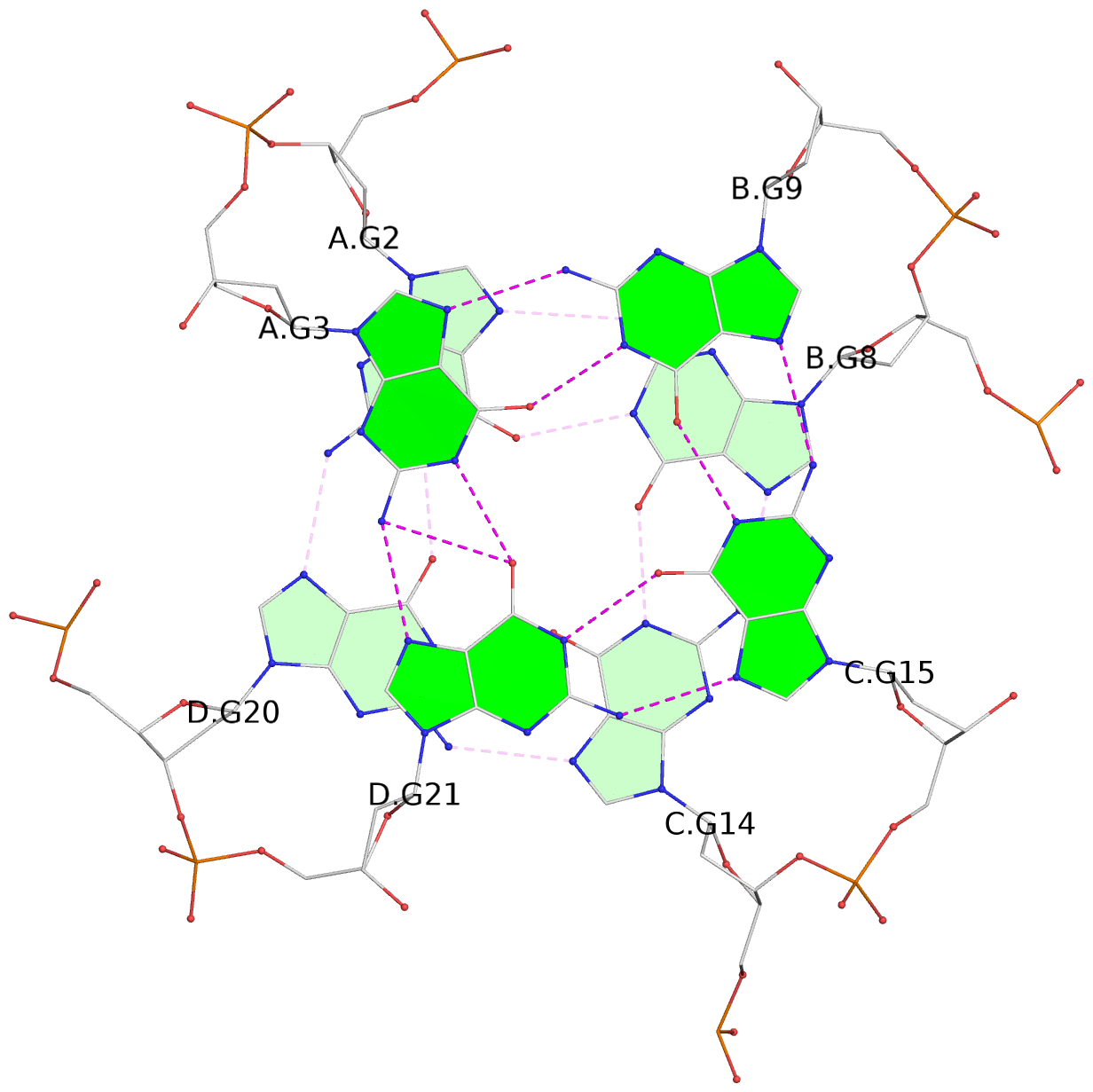
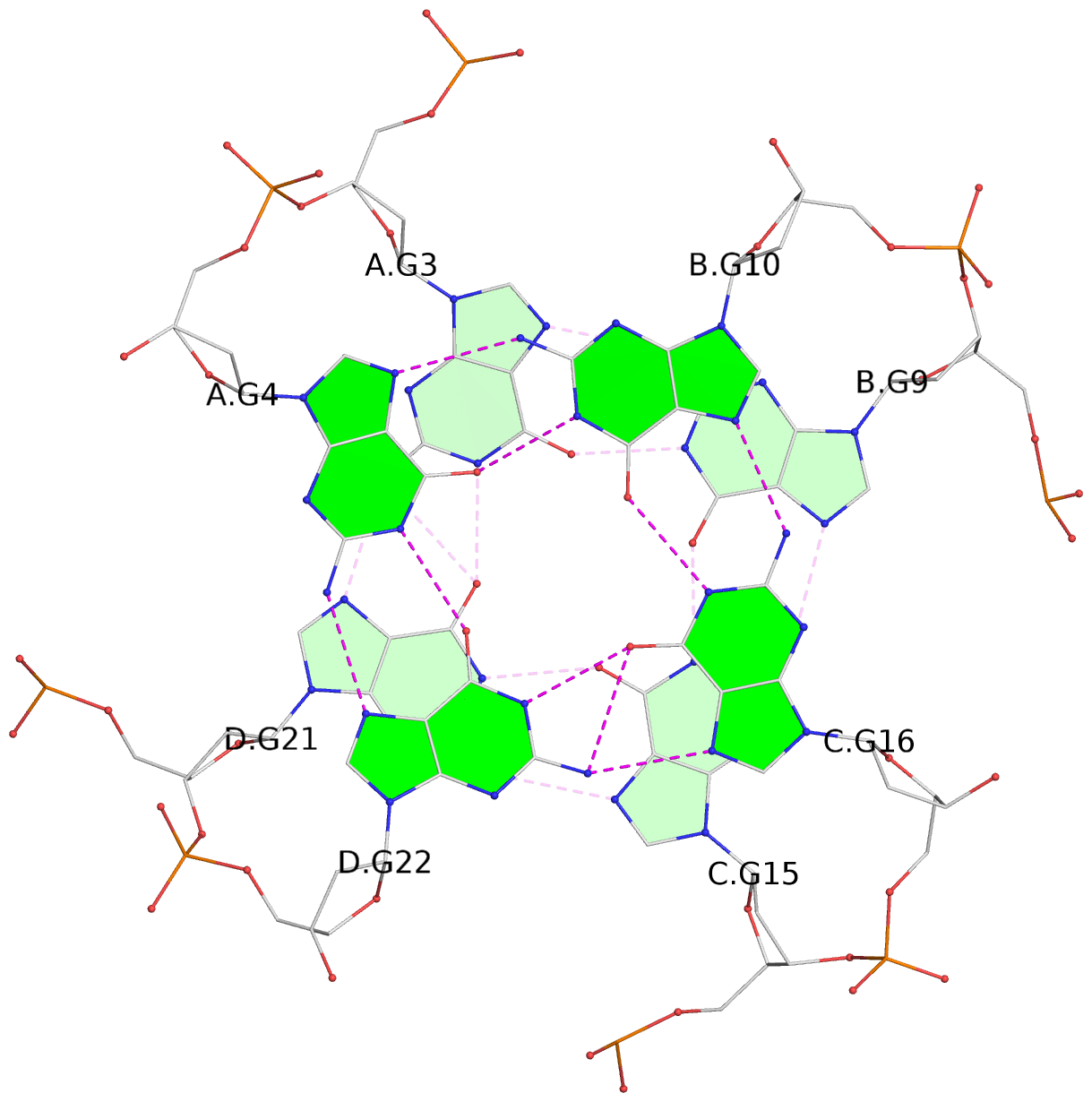
List of 4 G-tetrads
1 glyco-bond=ssss sugar=---- groove=---- planarity=0.122 type=planar nts=4 GGGG A.DG1,D.DG19,C.DG13,B.DG7 2 glyco-bond=---- sugar=---- groove=---- planarity=0.216 type=other nts=4 GGGG A.DG2,D.DG20,C.DG14,B.DG8 3 glyco-bond=---- sugar=---- groove=---- planarity=0.254 type=other nts=4 GGGG A.DG3,D.DG21,C.DG15,B.DG9 4 glyco-bond=---- sugar=---- groove=---- planarity=0.413 type=bowl nts=4 GGGG A.DG4,D.DG22,C.DG16,B.DG10
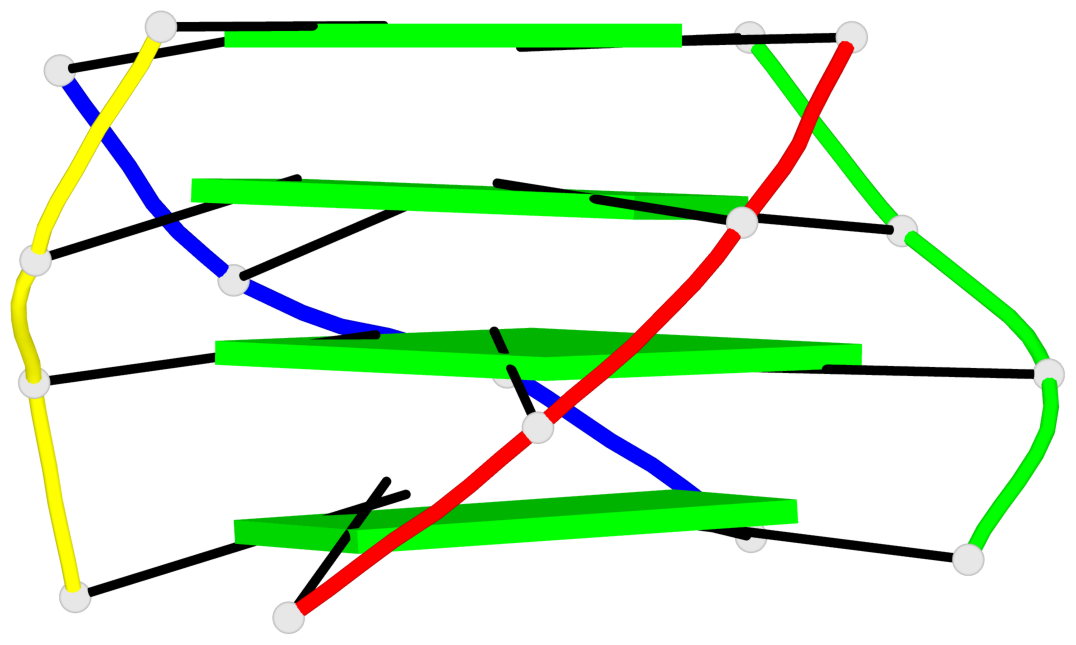
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.