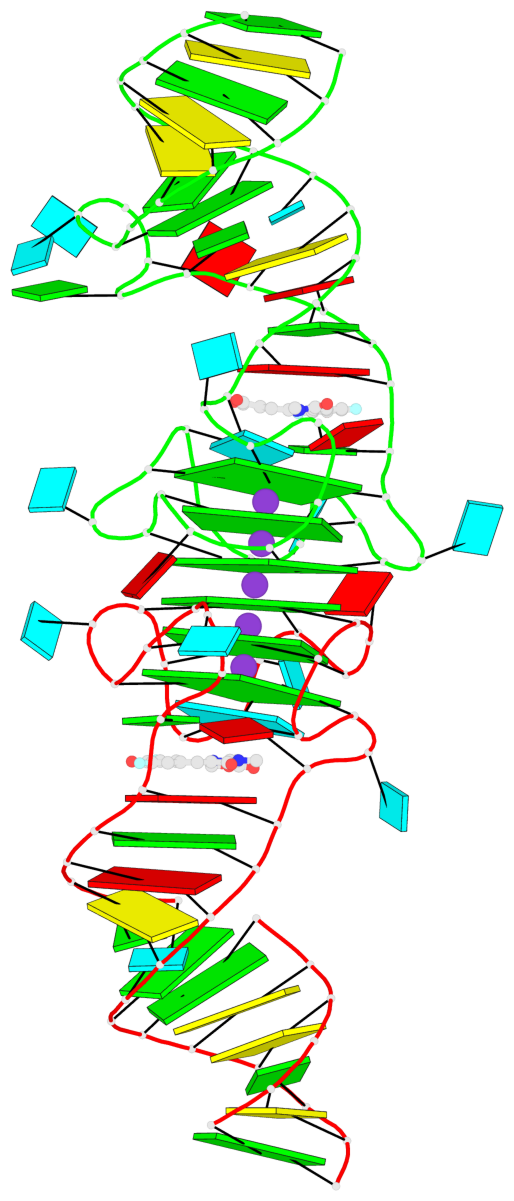
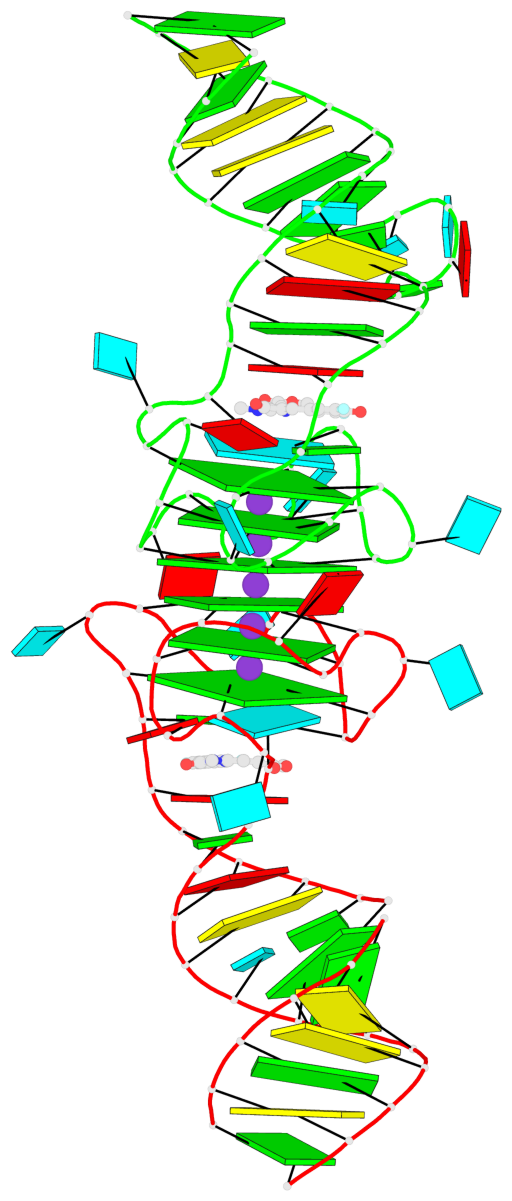
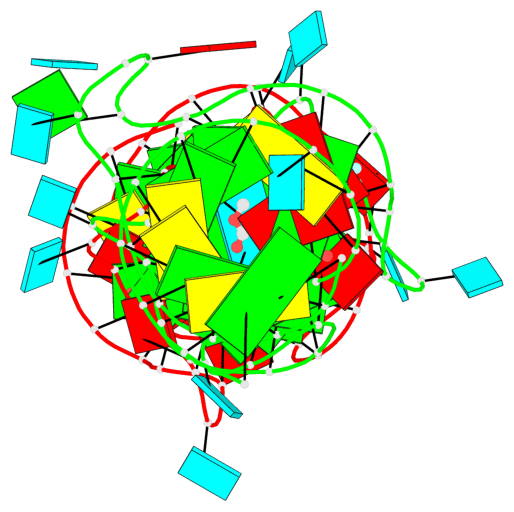
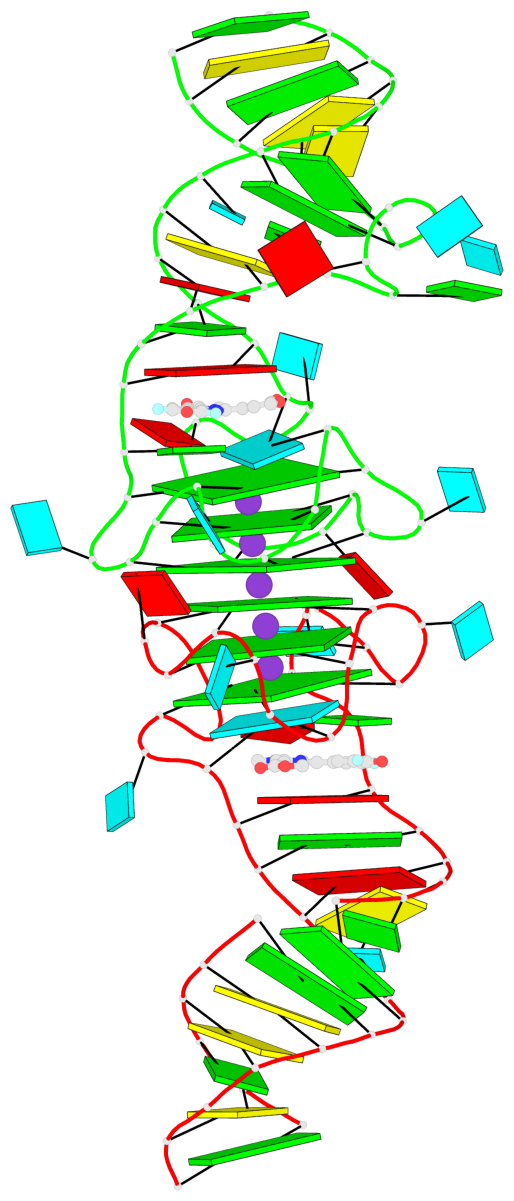
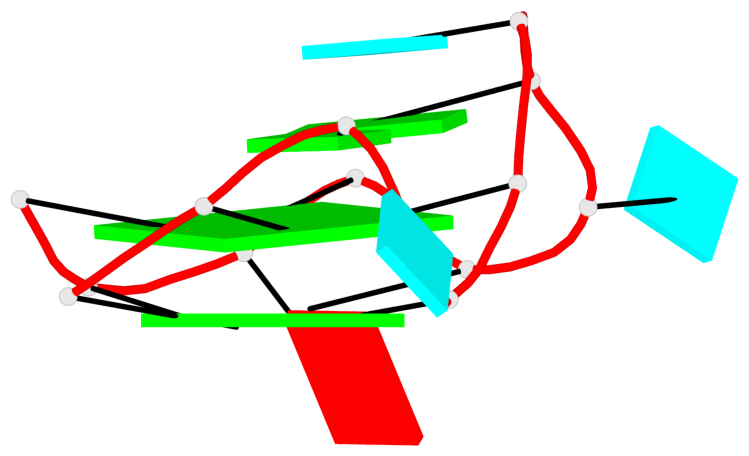
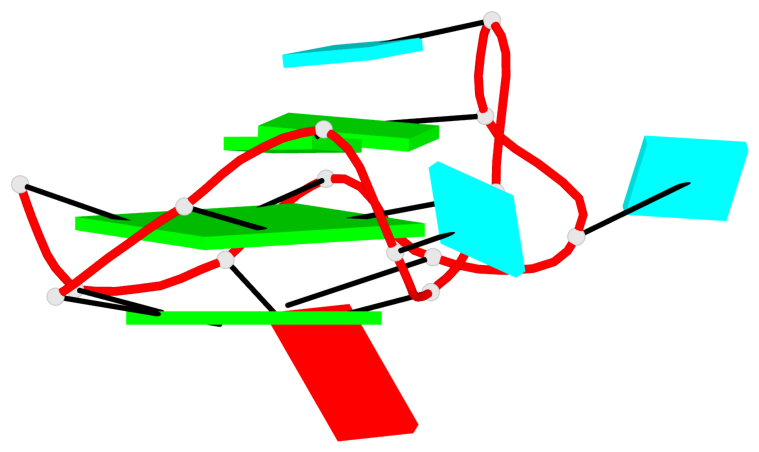
Detailed DSSR results for the G-quadruplex: PDB entry 8eyu
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8eyu
- Class
- RNA
- Method
- X-ray (1.95 Å)
- Summary
- Structure of beetroot dimer bound to dfame
- Reference
- Passalacqua LFM, Starich MR, Link KA, Wu J, Knutson JR, Tjandra N, Jaffrey SR, Ferre-D'Amare AR (2023): "Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture." Nat Commun, 14, 2969. doi: 10.1038/s41467-023-38683-3.
- Abstract
- Beetroot is a homodimeric in vitro selected RNA that binds and activates DFAME, a conditional fluorophore derived from GFP. It is 70% sequence-identical to the previously characterized homodimeric aptamer Corn, which binds one molecule of its cognate fluorophore DFHO at its interprotomer interface. We have now determined the Beetroot-DFAME co-crystal structure at 1.95 Å resolution, discovering that this RNA homodimer binds two molecules of the fluorophore, at sites separated by ~30 Å. In addition to this overall architectural difference, the local structures of the non-canonical, complex quadruplex cores of Beetroot and Corn are distinctly different, underscoring how subtle RNA sequence differences can give rise to unexpected structural divergence. Through structure-guided engineering, we generated a variant that has a 12-fold fluorescence activation selectivity switch toward DFHO. Beetroot and this variant form heterodimers and constitute the starting point for engineered tags whose through-space inter-fluorophore interaction could be used to monitor RNA dimerization.
- G4 notes
- 6 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
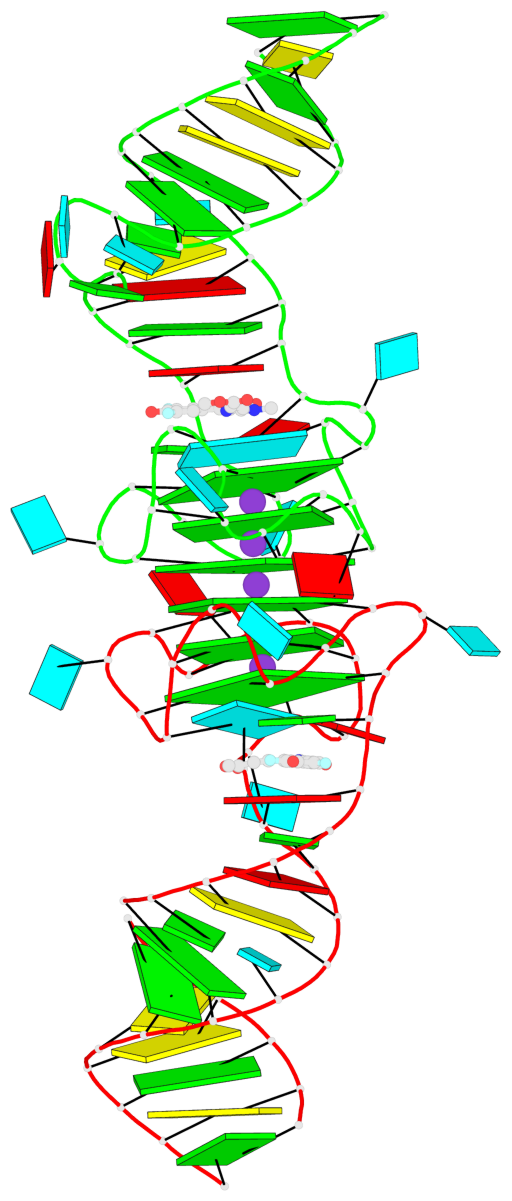
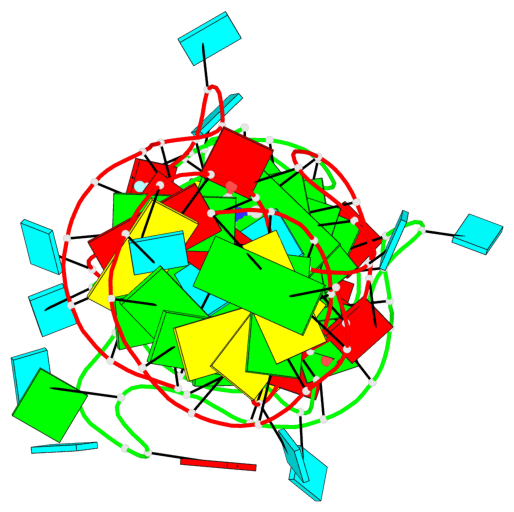
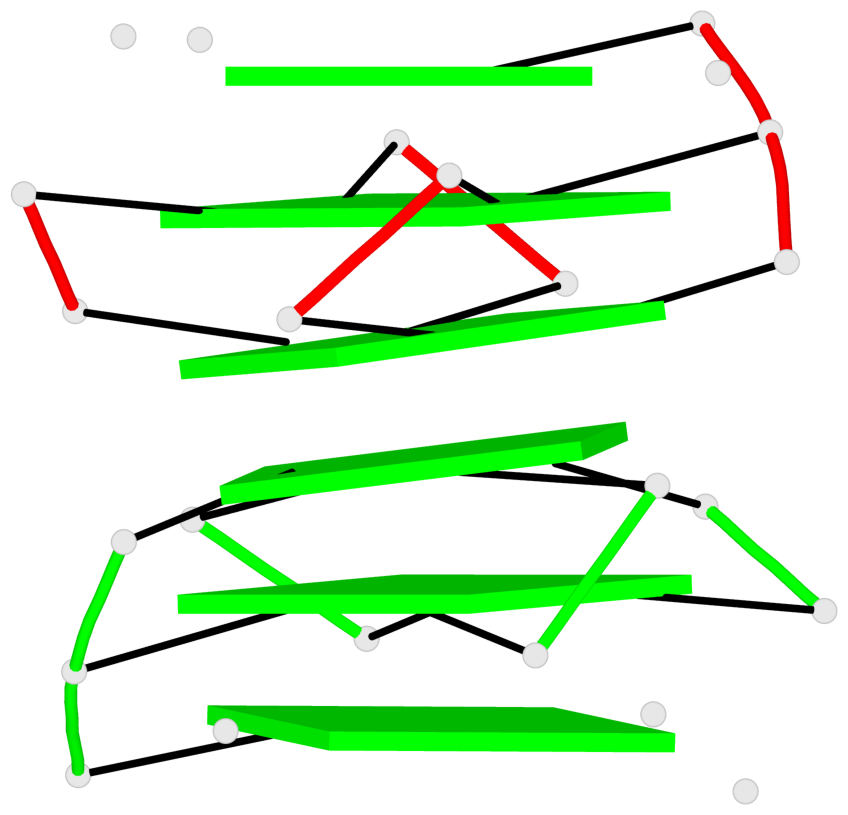
Base-block schematics in six views
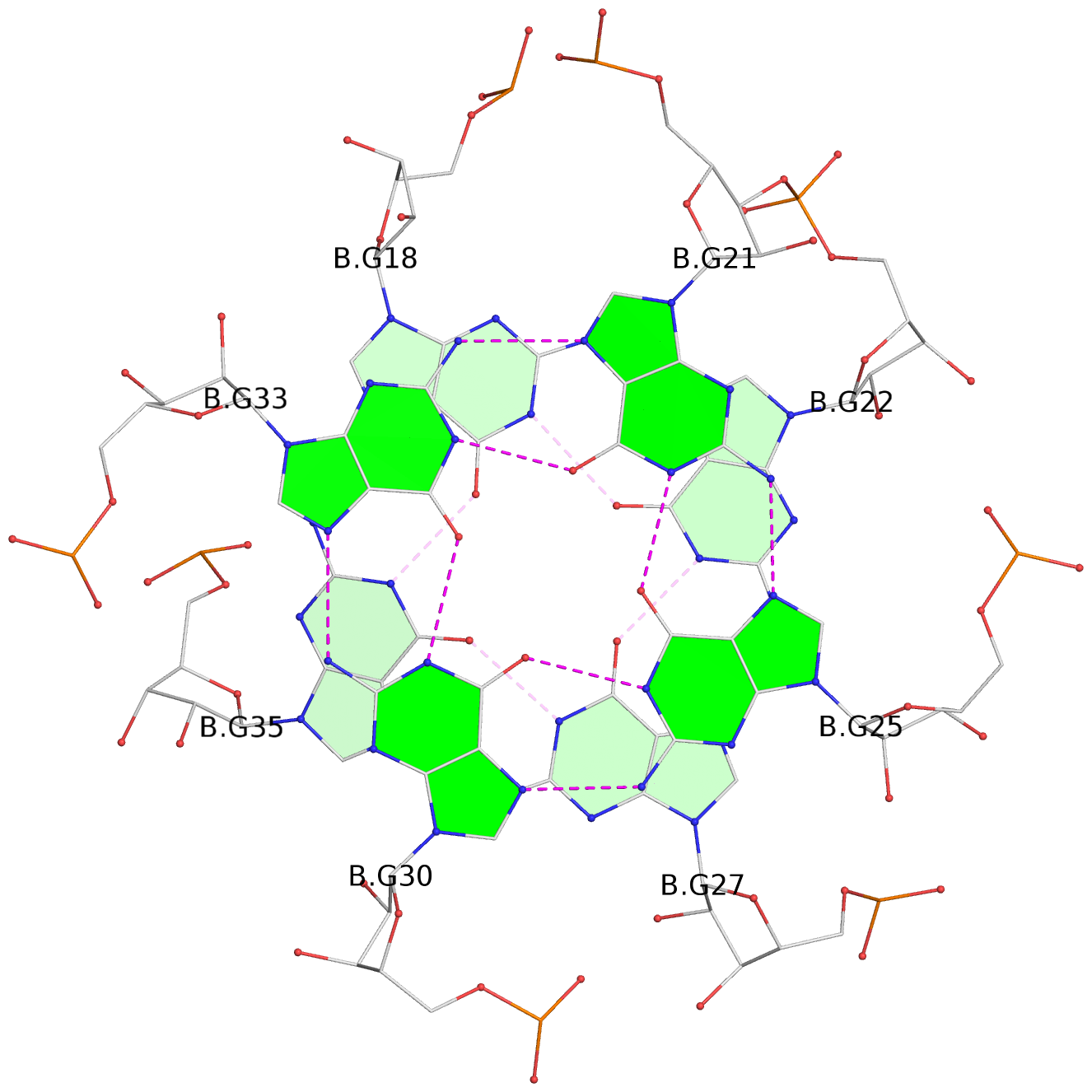
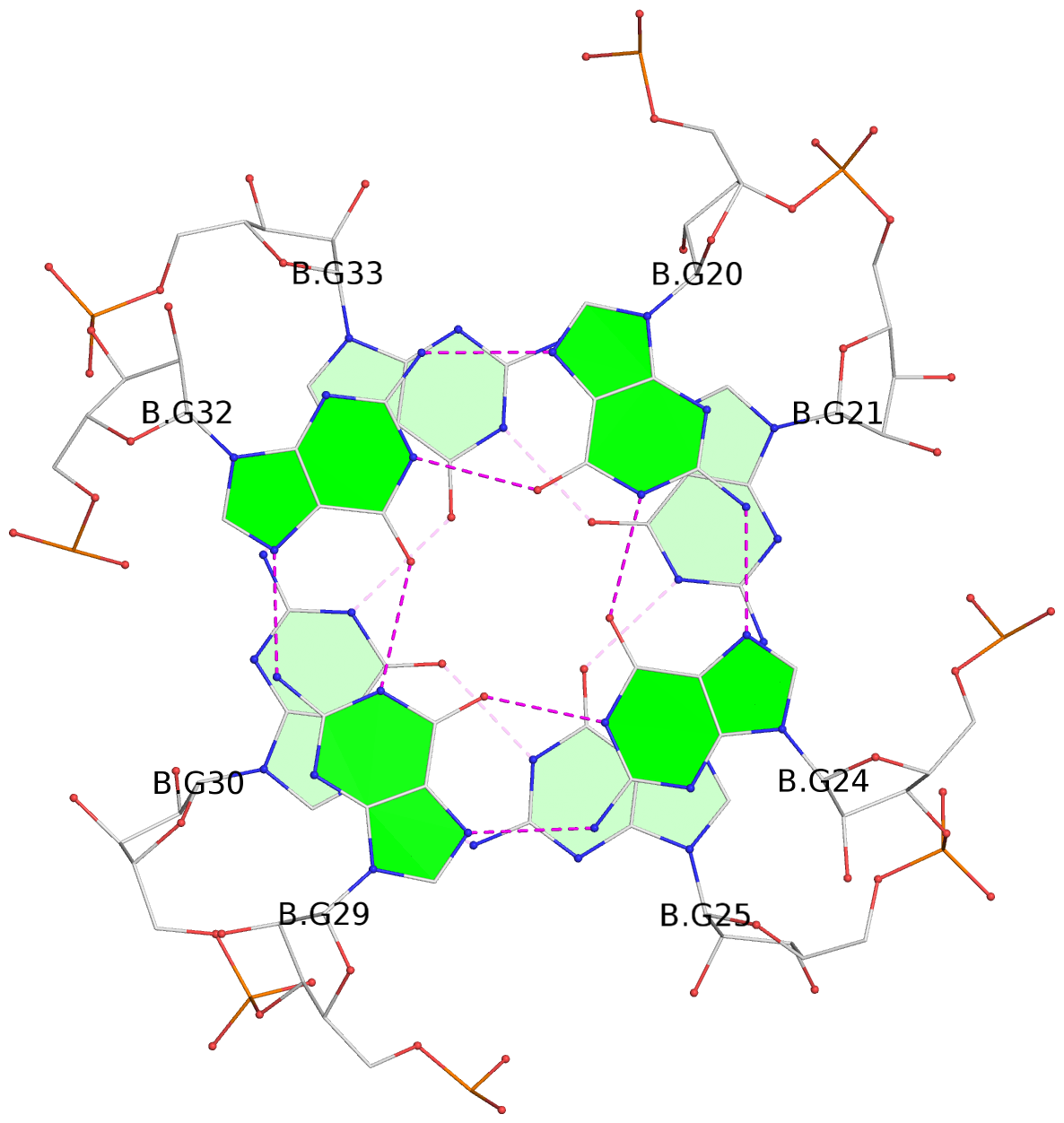
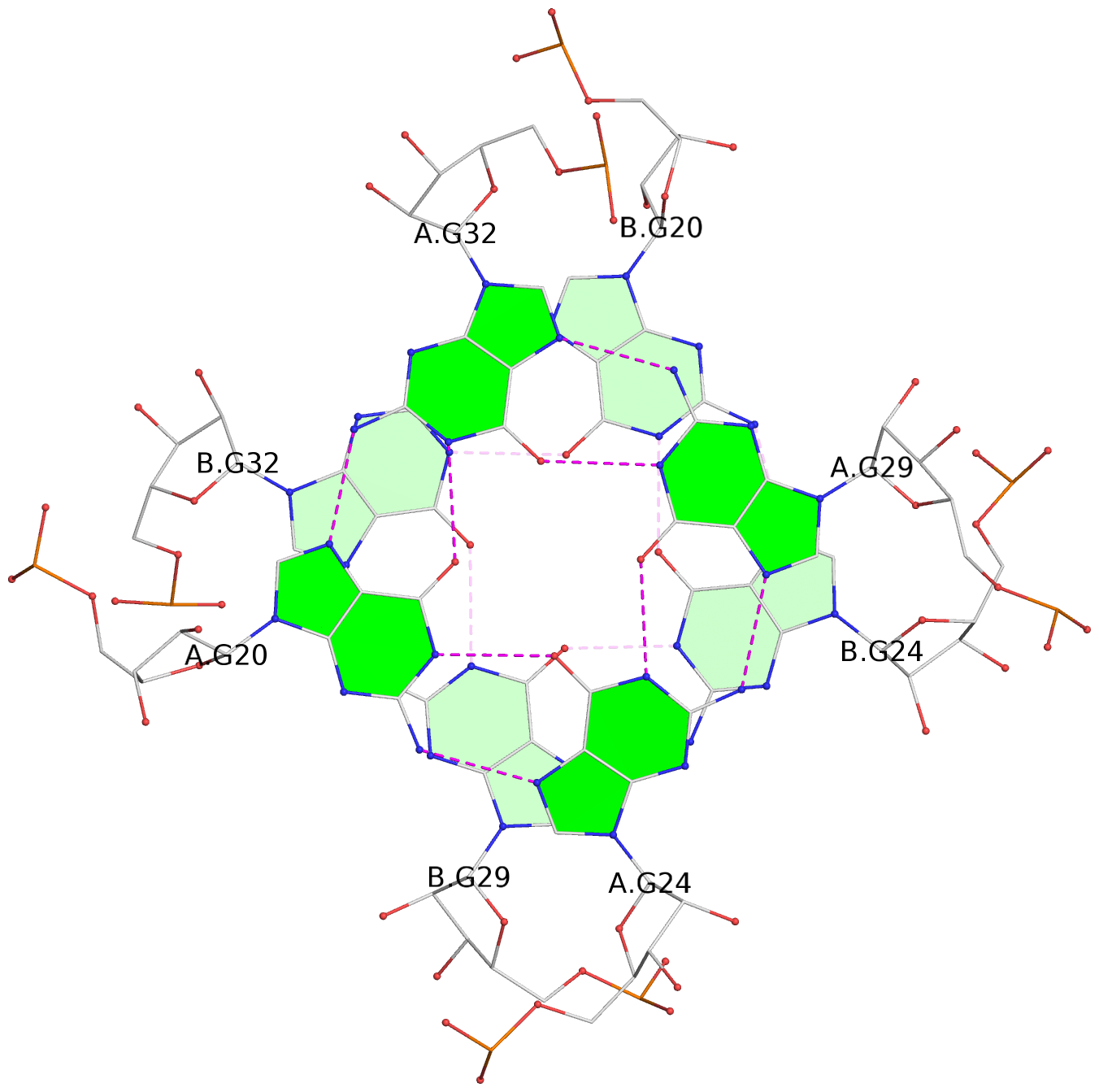
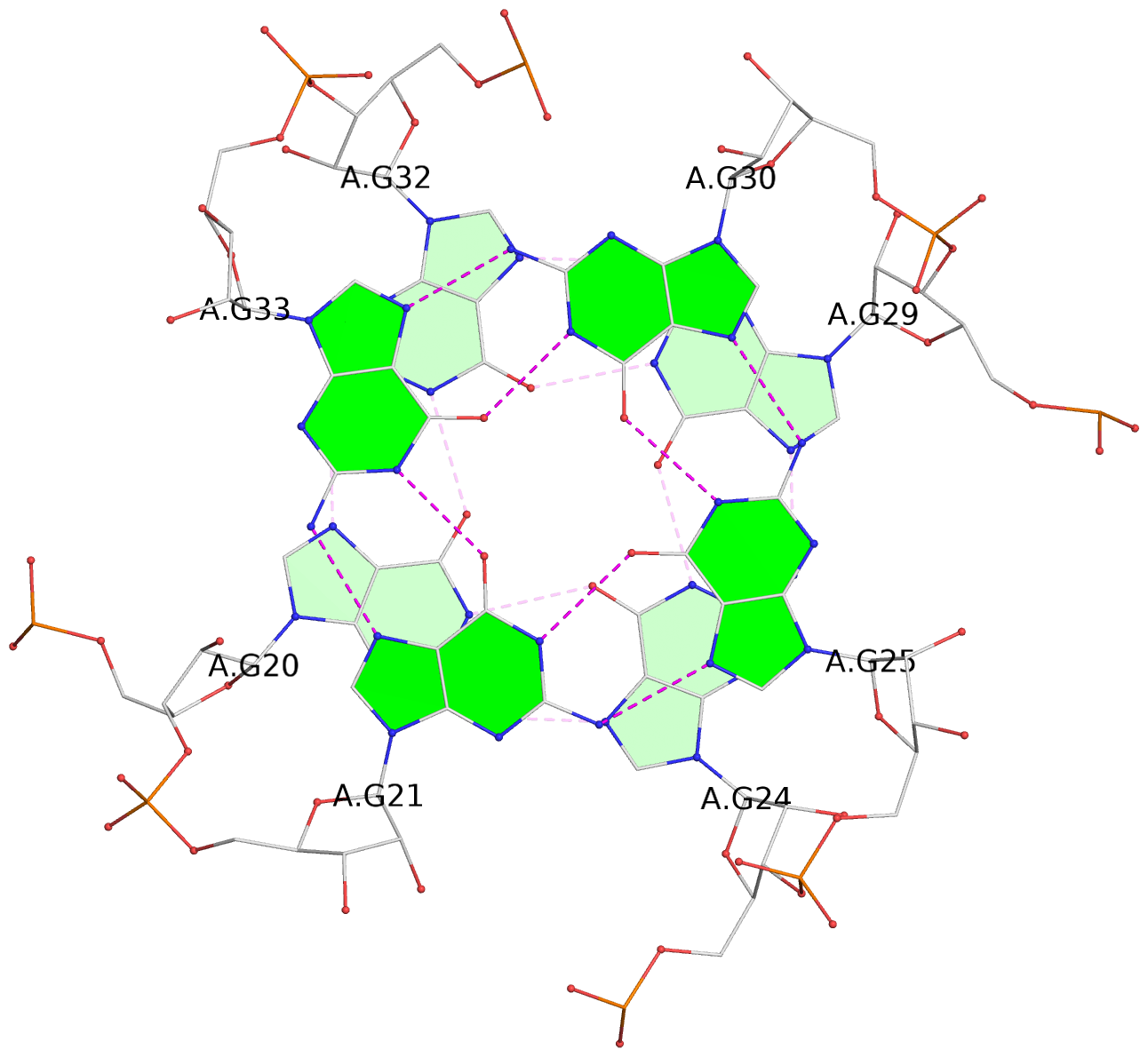
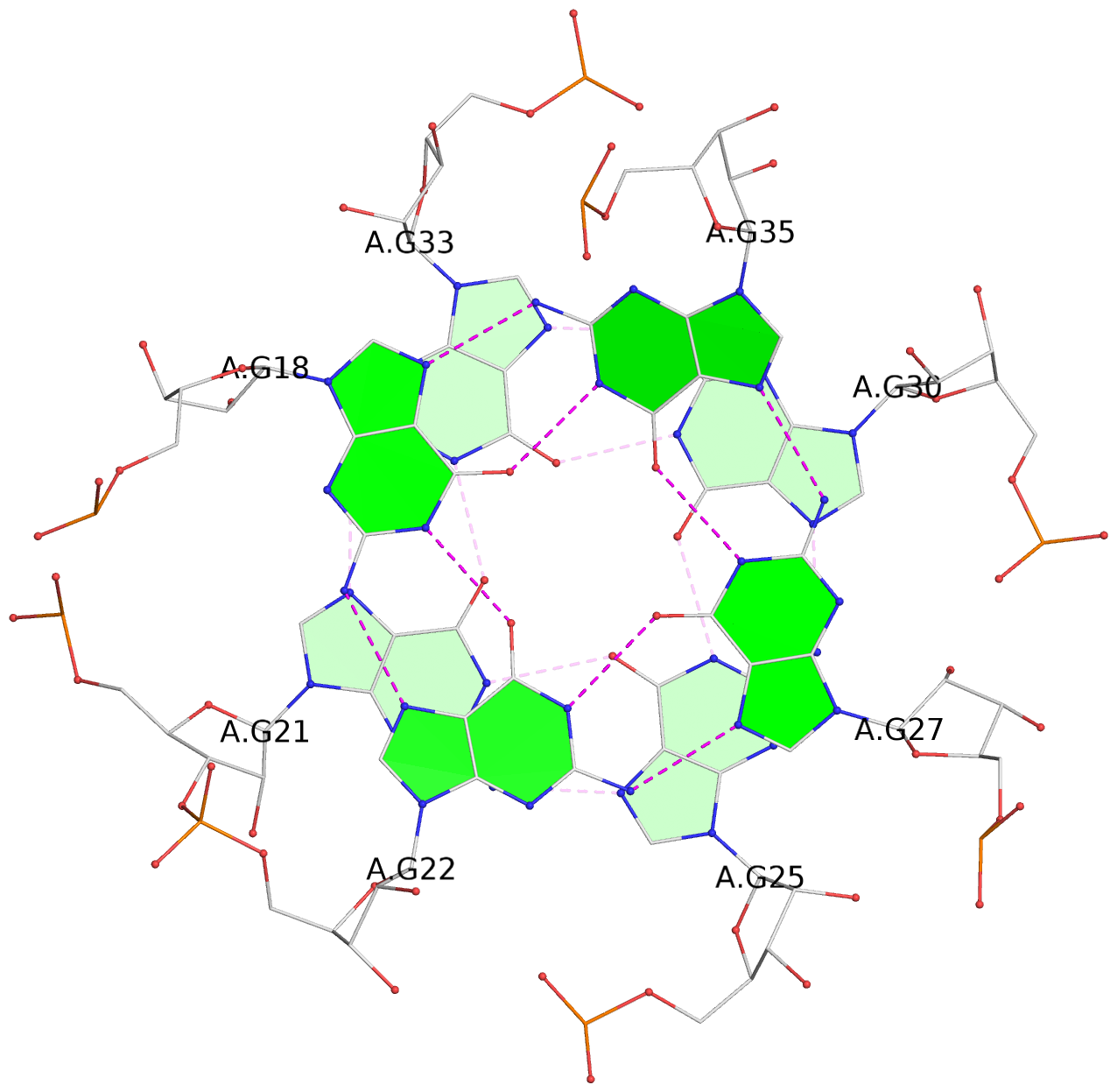
List of 6 G-tetrads
1 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.306 type=bowl nts=4 GGGG B.G18,B.G35,B.G27,B.G22 2 glyco-bond=---- sugar=-333 groove=---- planarity=0.252 type=other nts=4 GGGG B.G20,B.G24,B.G29,B.G32 3 glyco-bond=---- sugar=33-3 groove=---- planarity=0.128 type=planar nts=4 GGGG B.G21,B.G25,B.G30,B.G33 4 glyco-bond=sss- sugar=---- groove=--wn planarity=0.352 type=bowl-2 nts=4 GGGG A.G18,A.G35,A.G27,A.G22 5 glyco-bond=---- sugar=-333 groove=---- planarity=0.281 type=other nts=4 GGGG A.G20,A.G24,A.G29,A.G32 6 glyco-bond=---- sugar=33-3 groove=---- planarity=0.152 type=planar nts=4 GGGG A.G21,A.G25,A.G30,A.G33
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']