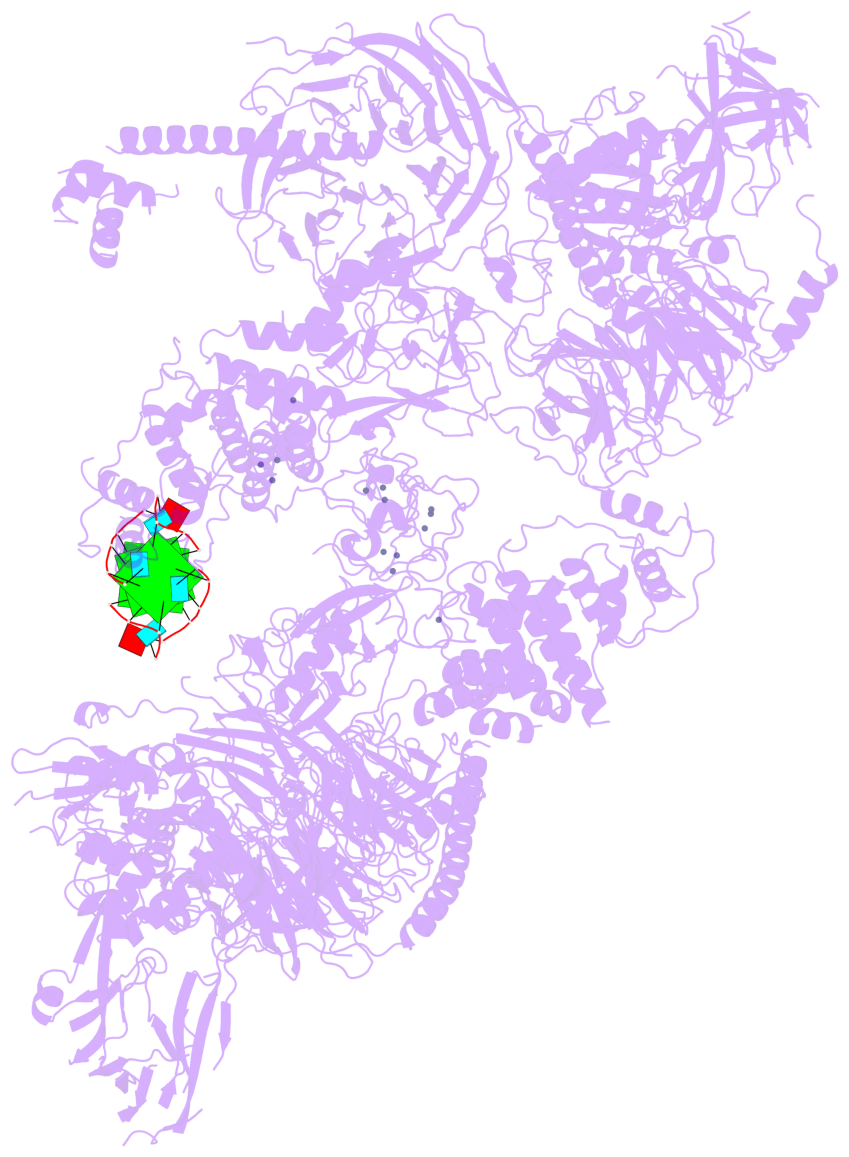
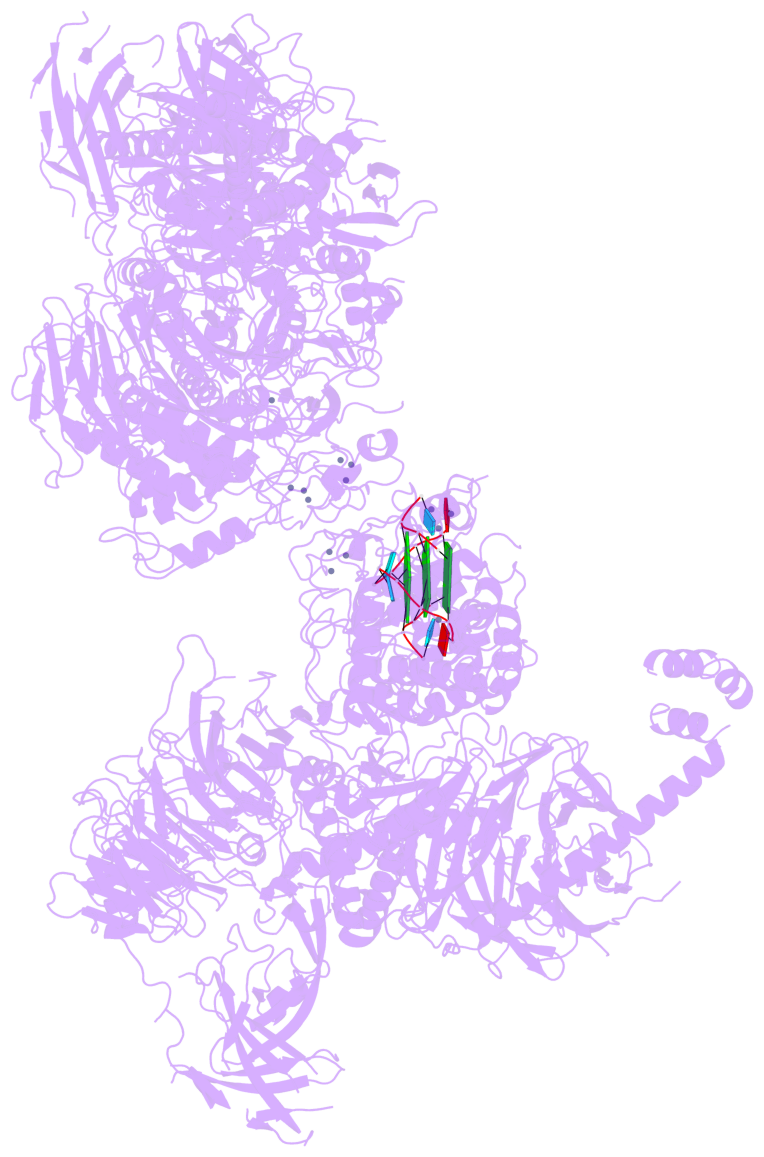
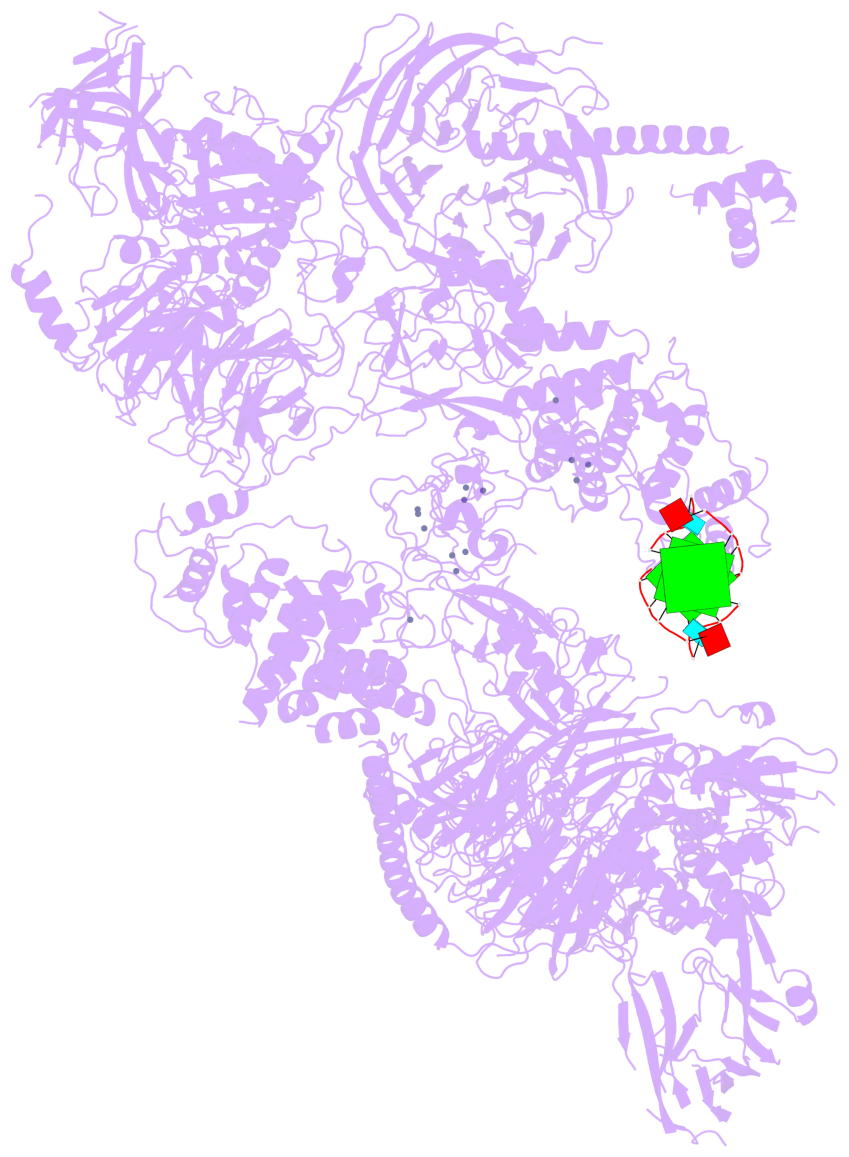
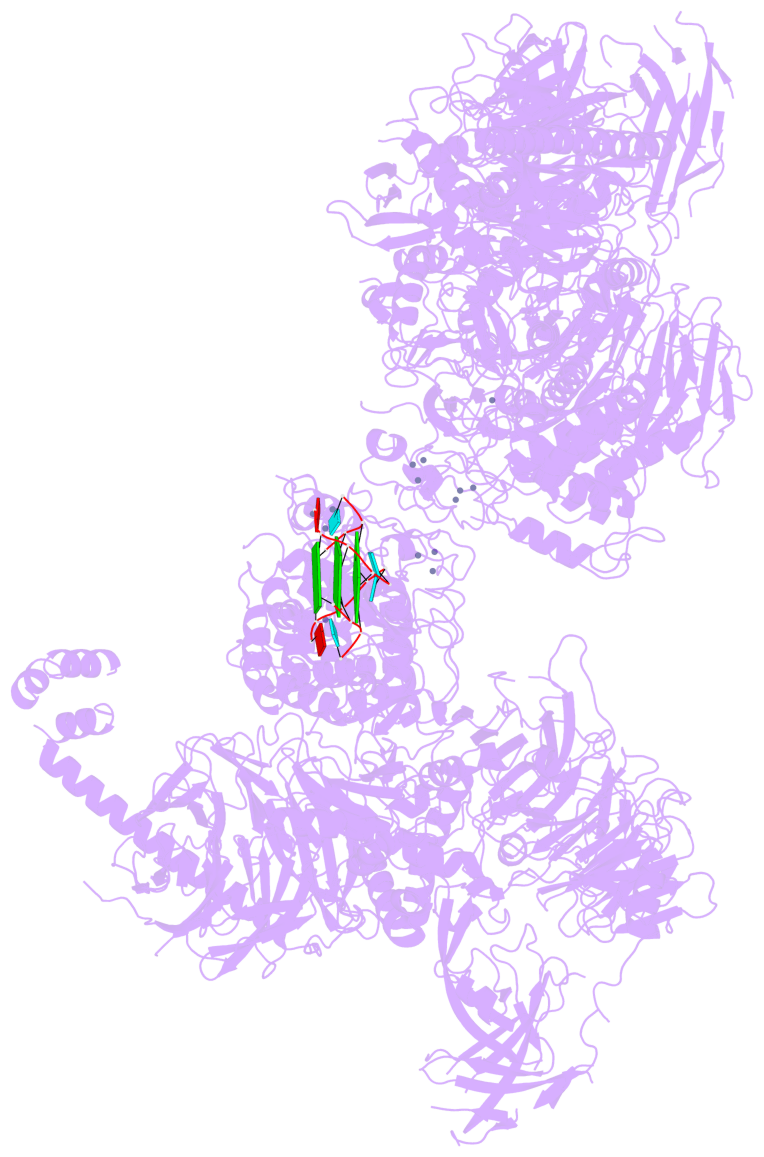
Detailed DSSR results for the G-quadruplex: PDB entry 8fyh
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8fyh
- Class
- gene regulation
- Method
- cryo-EM (3.4 Å)
- Summary
- G4 RNA-mediated prc2 dimer
- Reference
- Song J, Gooding AR, Hemphill WO, Love BD, Robertson A, Yao L, Zon LI, North TE, Kasinath V, Cech TR (2023): "Structural basis for inactivation of PRC2 by G-quadruplex RNA." Science, 381, 1331-1337. doi: 10.1126/science.adh0059.
- Abstract
- Polycomb repressive complex 2 (PRC2) silences genes through trimethylation of histone H3K27. PRC2 associates with numerous precursor messenger RNAs (pre-mRNAs) and long noncoding RNAs (lncRNAs) with a binding preference for G-quadruplex RNA. In this work, we present a 3.3-Å-resolution cryo-electron microscopy structure of PRC2 bound to a G-quadruplex RNA. Notably, RNA mediates the dimerization of PRC2 by binding both protomers and inducing a protein interface composed of two copies of the catalytic subunit EZH2, thereby blocking nucleosome DNA interaction and histone H3 tail accessibility. Furthermore, an RNA-binding loop of EZH2 facilitates the handoff between RNA and DNA, another activity implicated in PRC2 regulation by RNA. We identified a gain-of-function mutation in this loop that activates PRC2 in zebrafish. Our results reveal mechanisms for RNA-mediated regulation of a chromatin-modifying enzyme.
- G4 notes
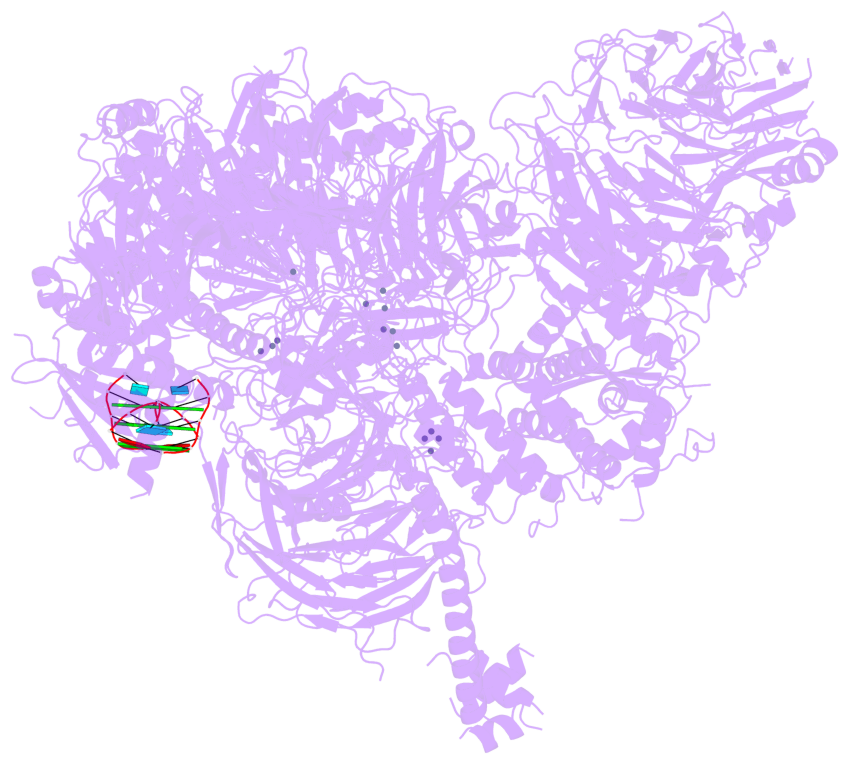
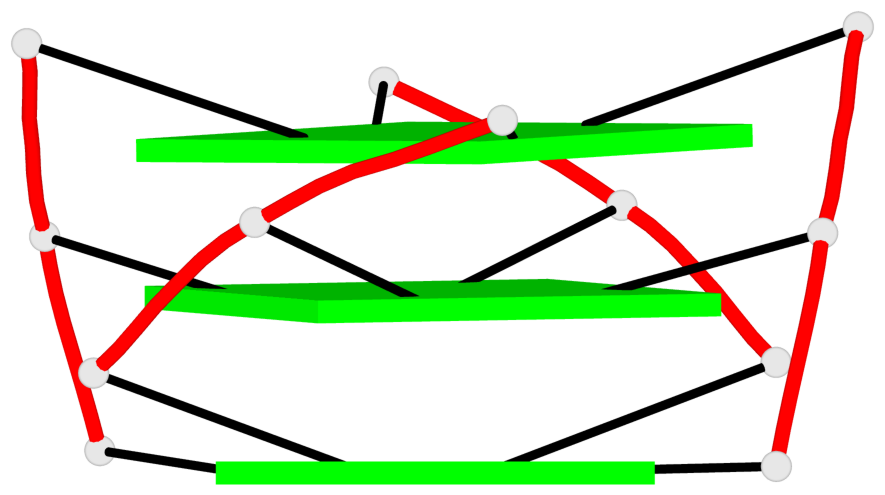
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, , parallel(4+0), UUUU
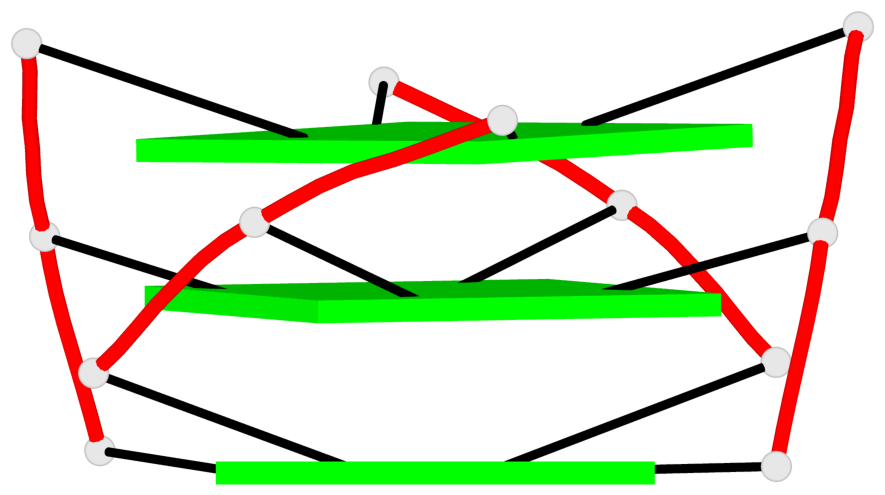
Base-block schematics in six views
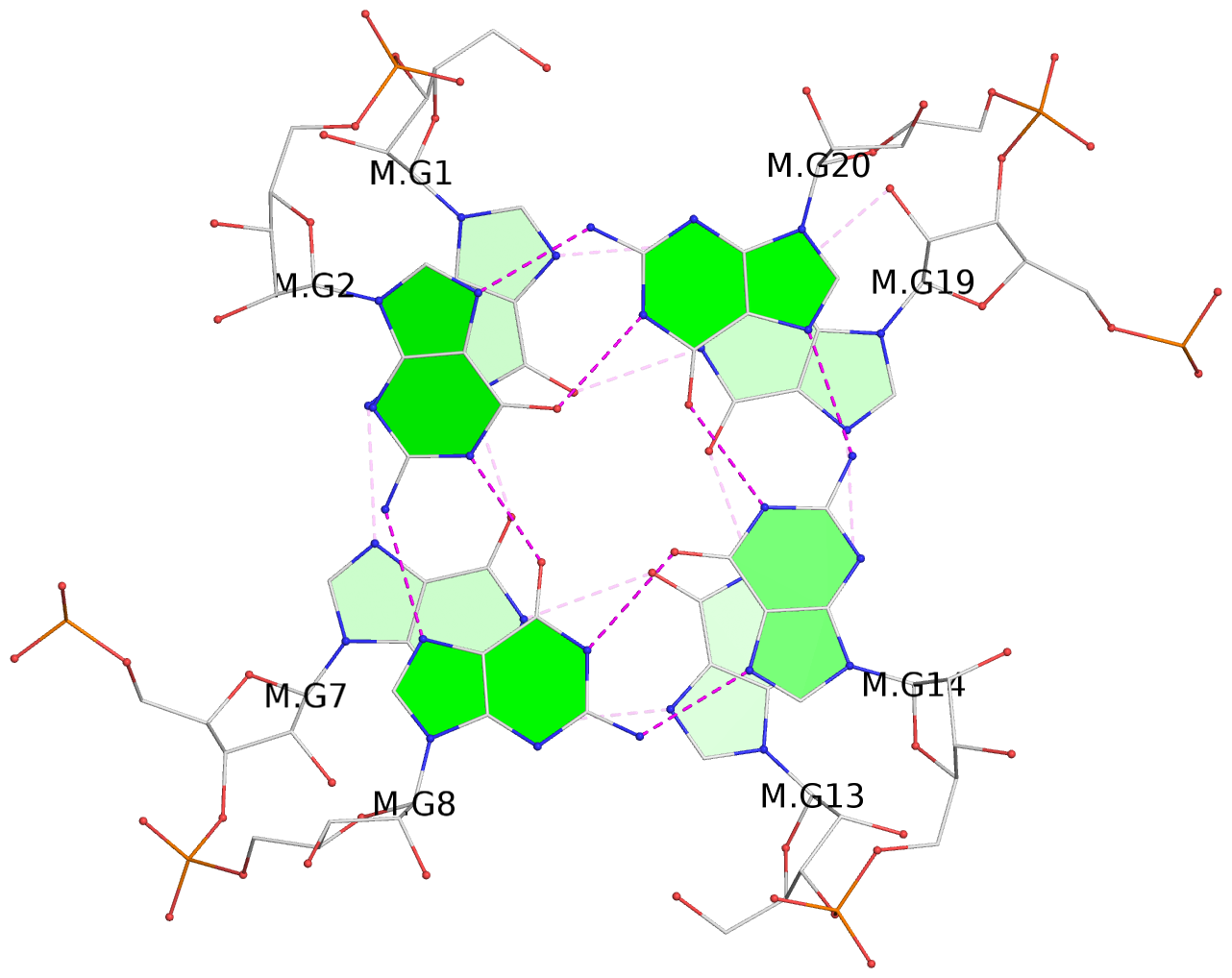
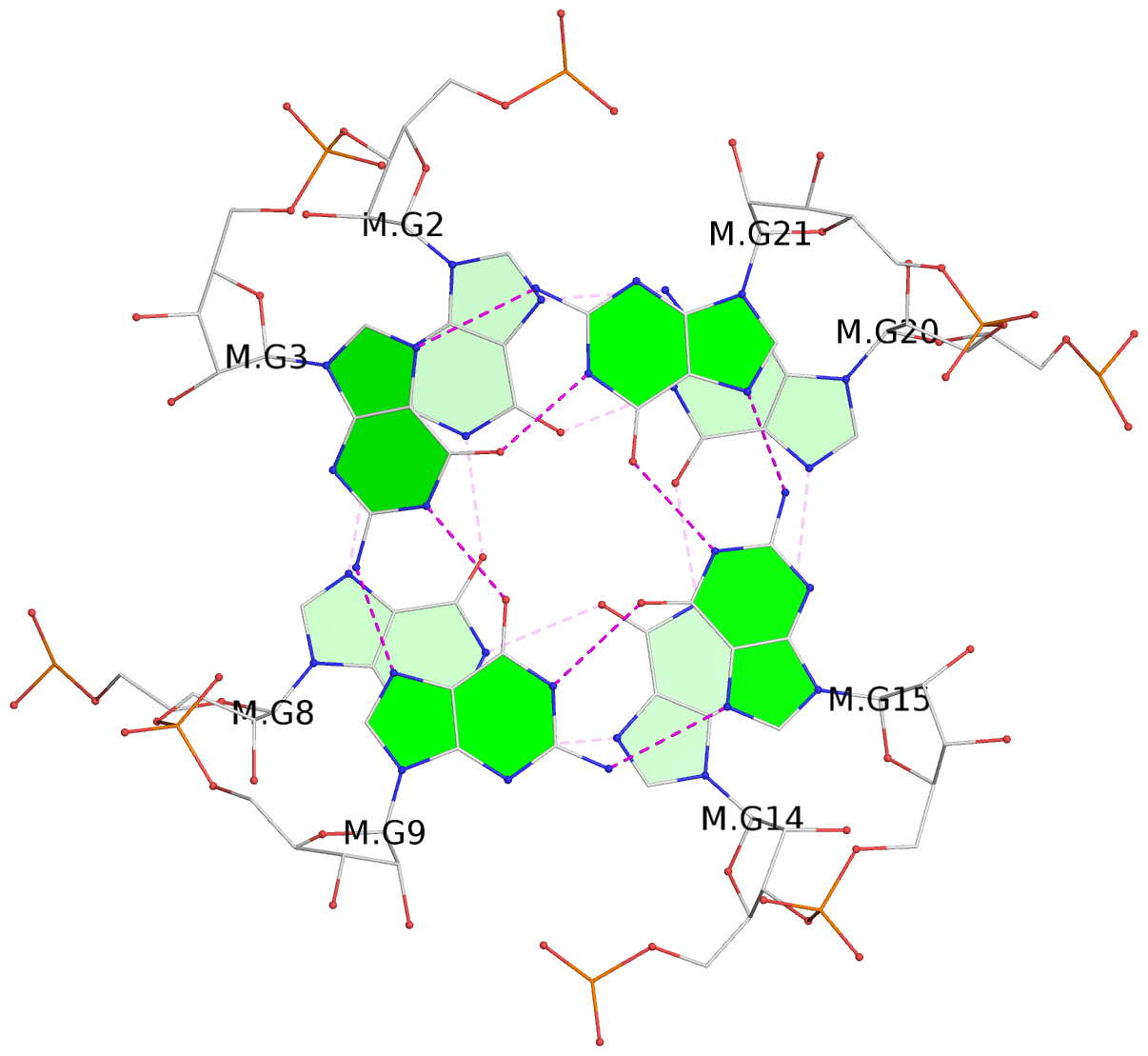
List of 3 G-tetrads
1 glyco-bond=---- sugar=3.3. groove=---- planarity=0.494 type=other nts=4 GGGG M.G1,M.G7,M.G13,M.G19 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.419 type=saddle nts=4 GGGG M.G2,M.G8,M.G14,M.G20 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.433 type=other nts=4 GGGG M.G3,M.G9,M.G15,M.G21
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.