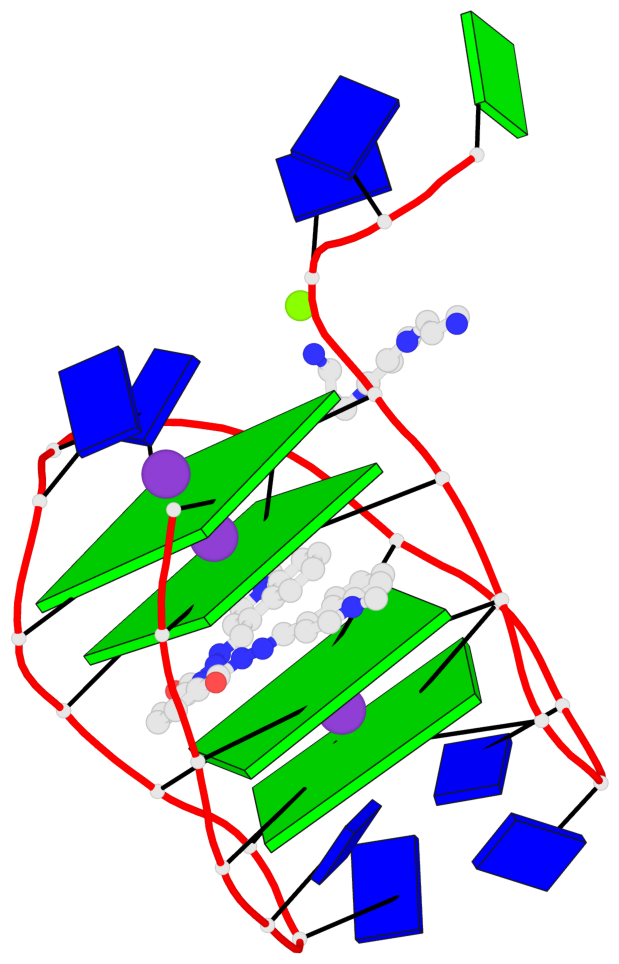
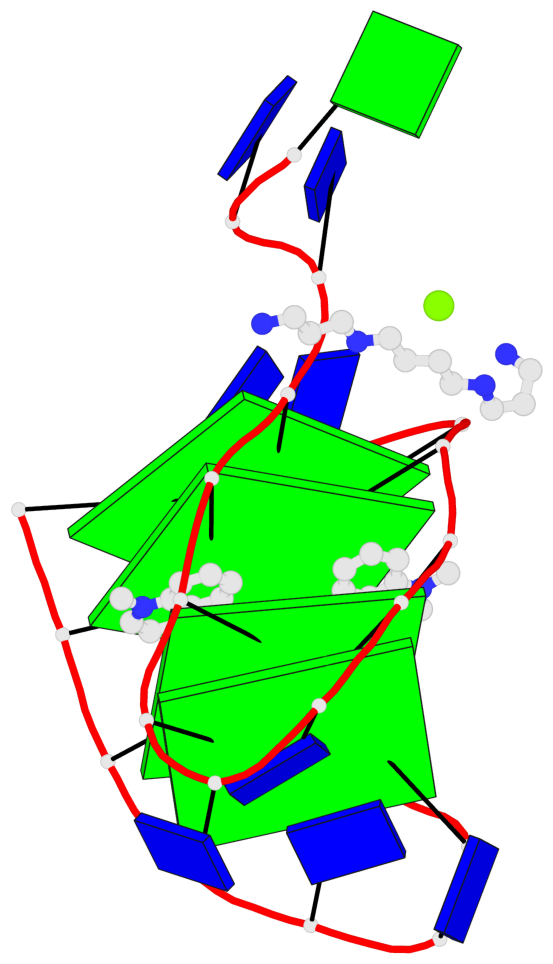
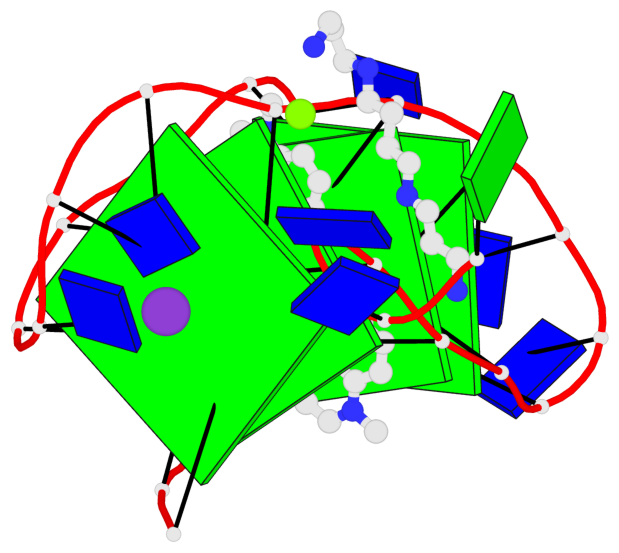
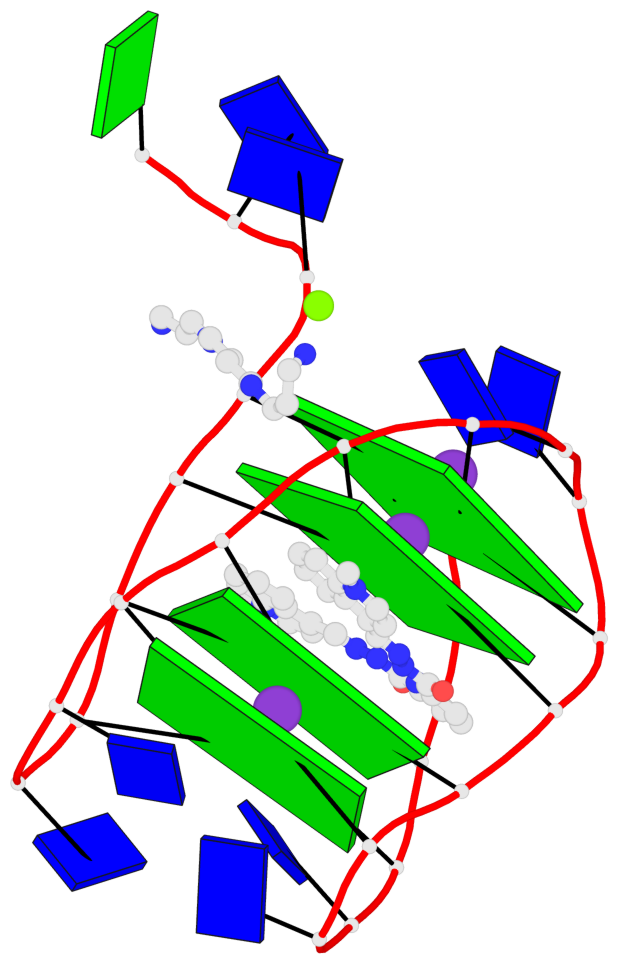
Detailed DSSR results for the G-quadruplex: PDB entry 8g2n
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8g2n
- Class
- DNA
- Method
- X-ray (1.33 Å)
- Summary
- Crystal structure of tetrahymena thermophila g-rich DNA with novel ligand pydh2
- Reference
- Martin KN, Yatsunyk LA: "Crystal structure of Tetrahymena thermophila G-rich DNA with novel ligand PyDH2."
- Abstract
- G4 notes
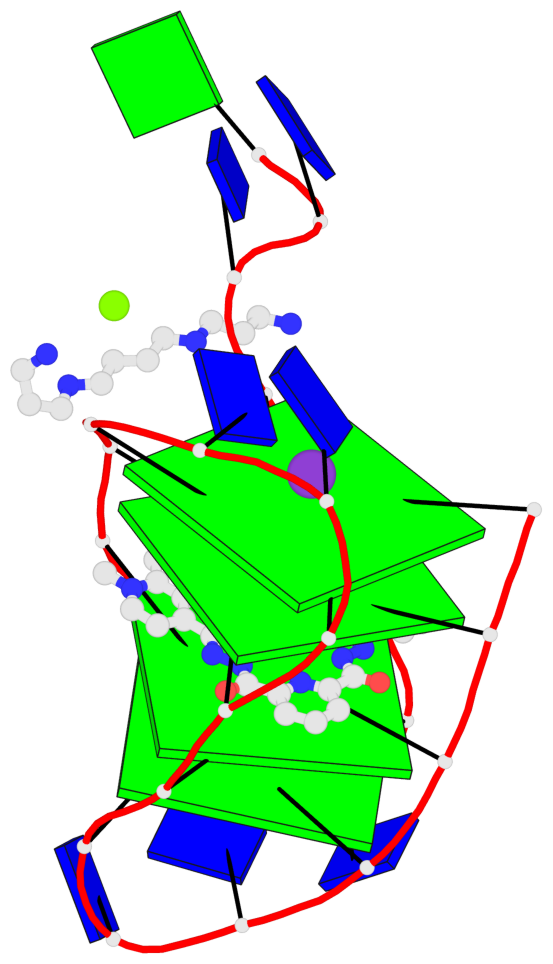
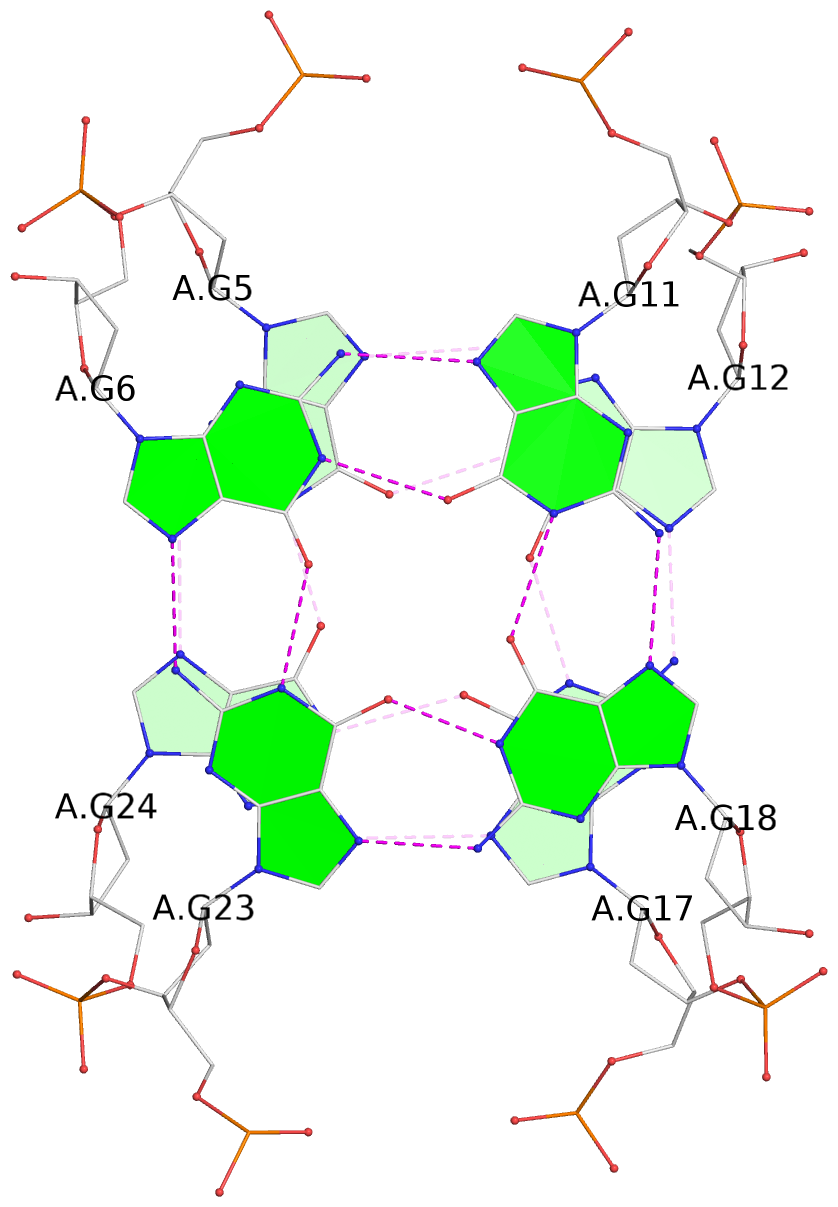
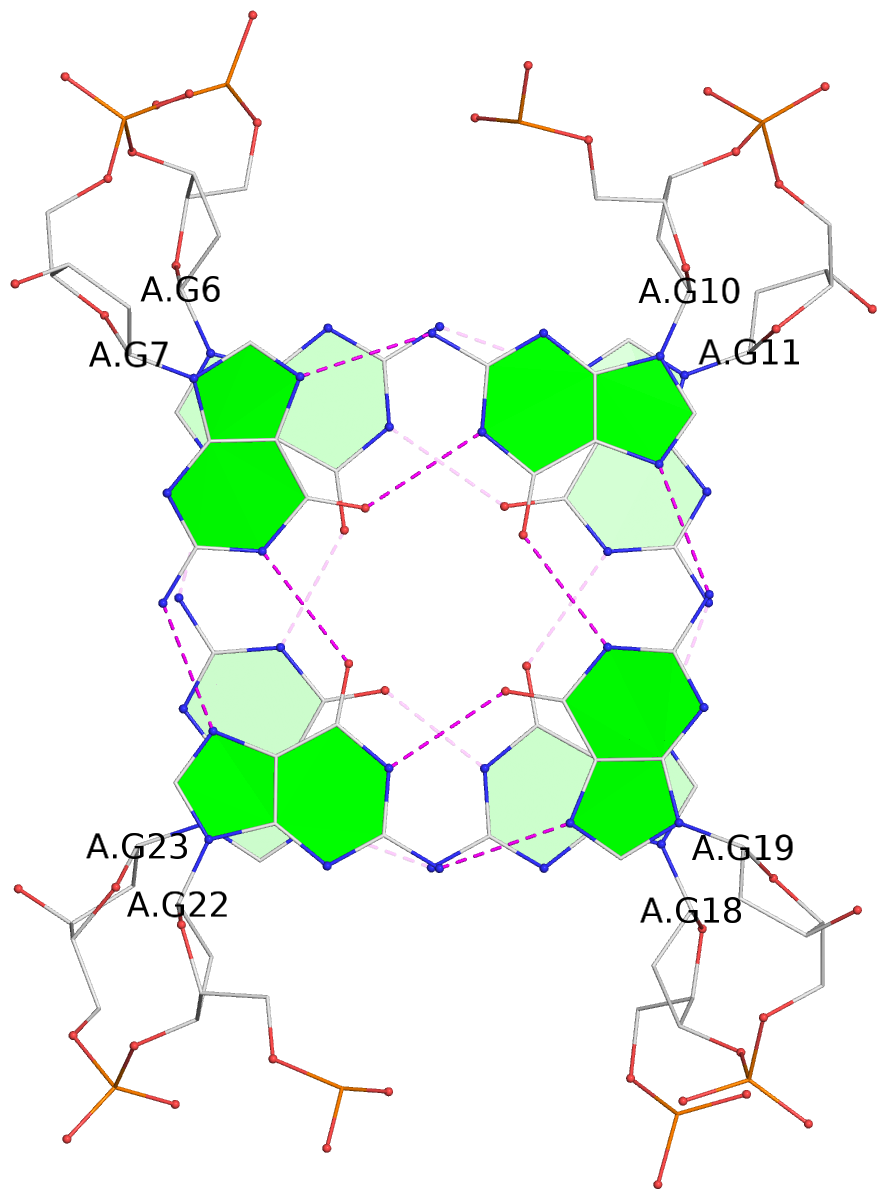
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, 4(+Ln+Lw+Ln), chair(2+2), UDUD
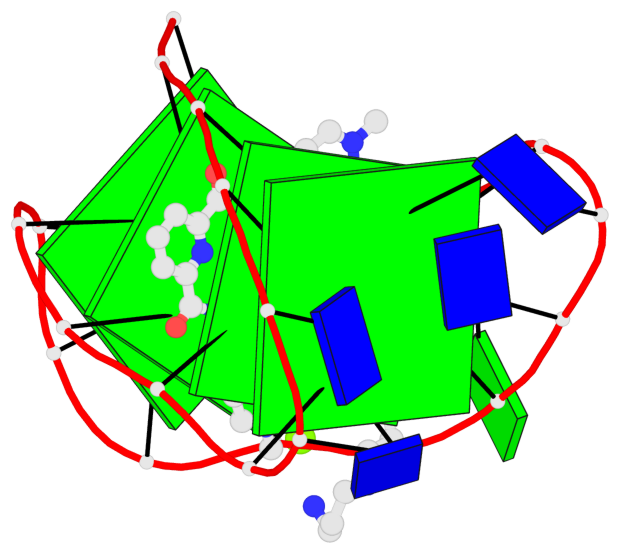
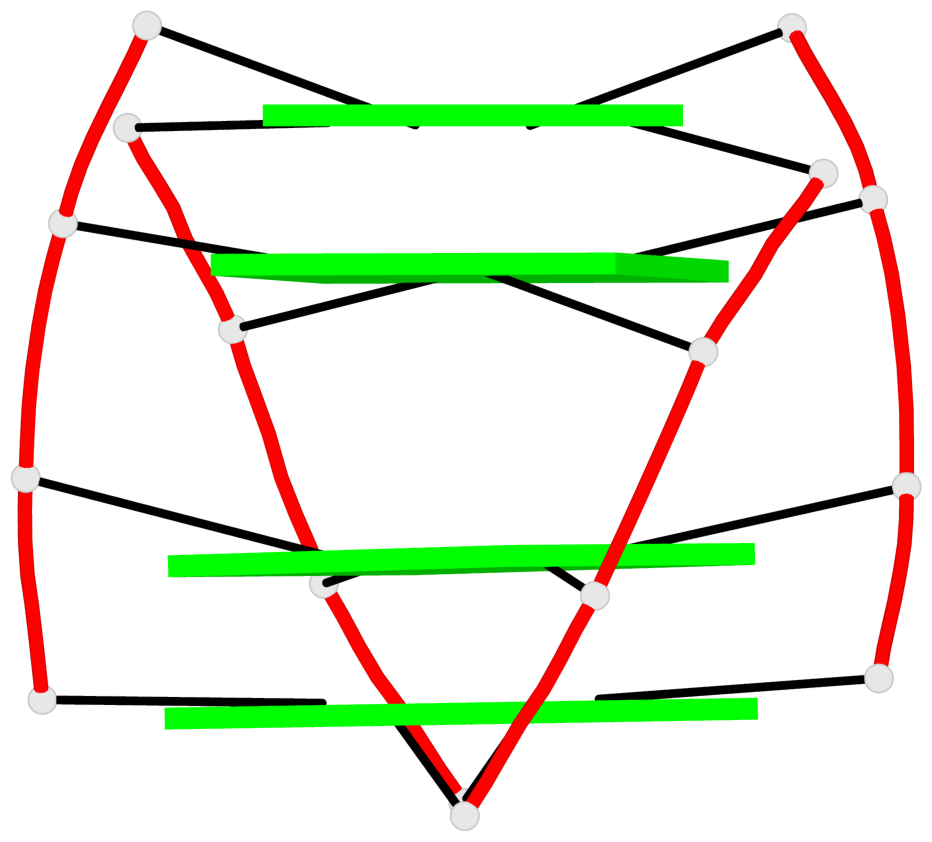
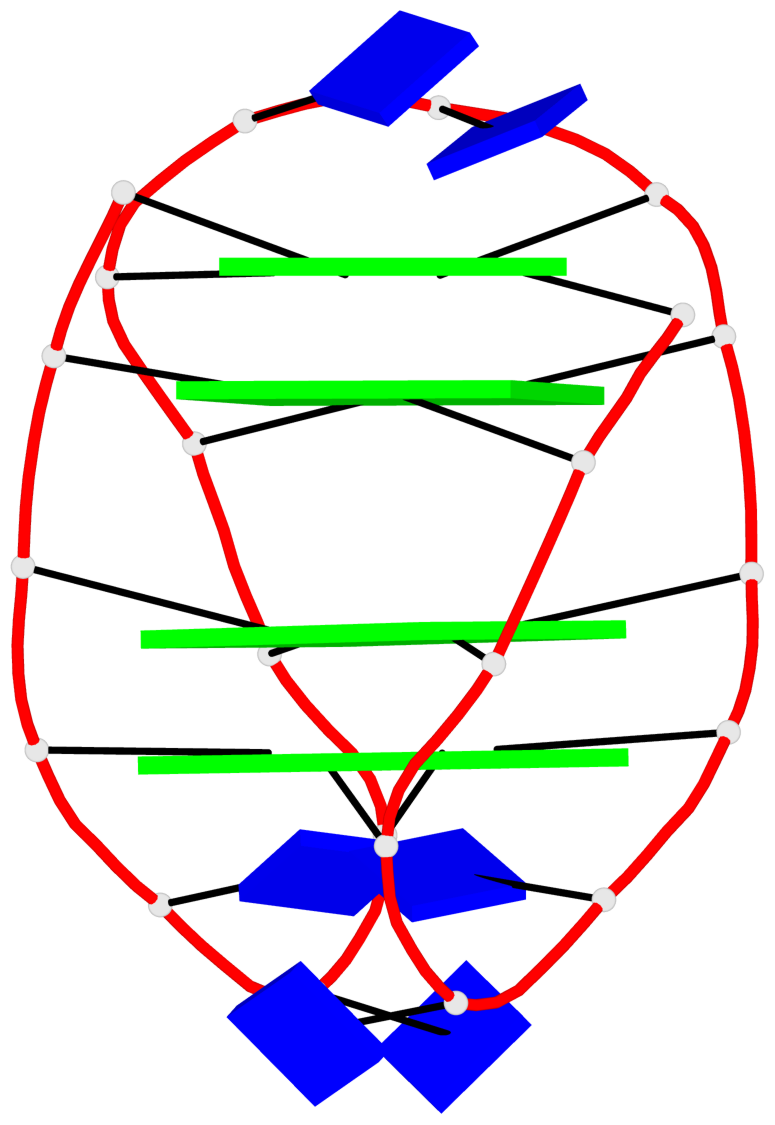
Base-block schematics in six views
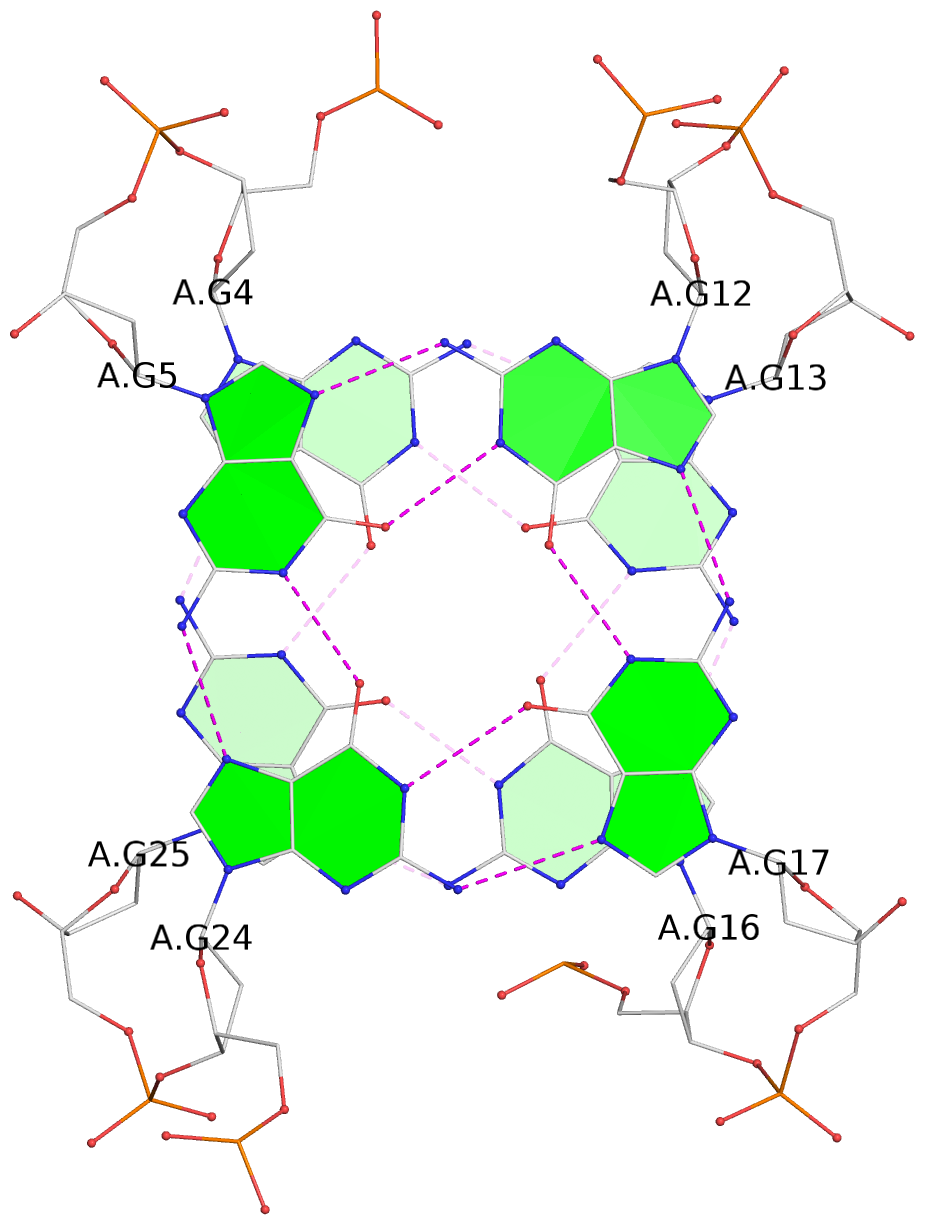
List of 4 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.543 type=other nts=4 GGGG A.DG4,A.DG25,A.DG16,A.DG13 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.529 type=other nts=4 GGGG A.DG5,A.DG24,A.DG17,A.DG12 3 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.447 type=other nts=4 GGGG A.DG6,A.DG23,A.DG18,A.DG11 4 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.522 type=bowl-2 nts=4 GGGG A.DG7,A.DG22,A.DG19,A.DG10
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.