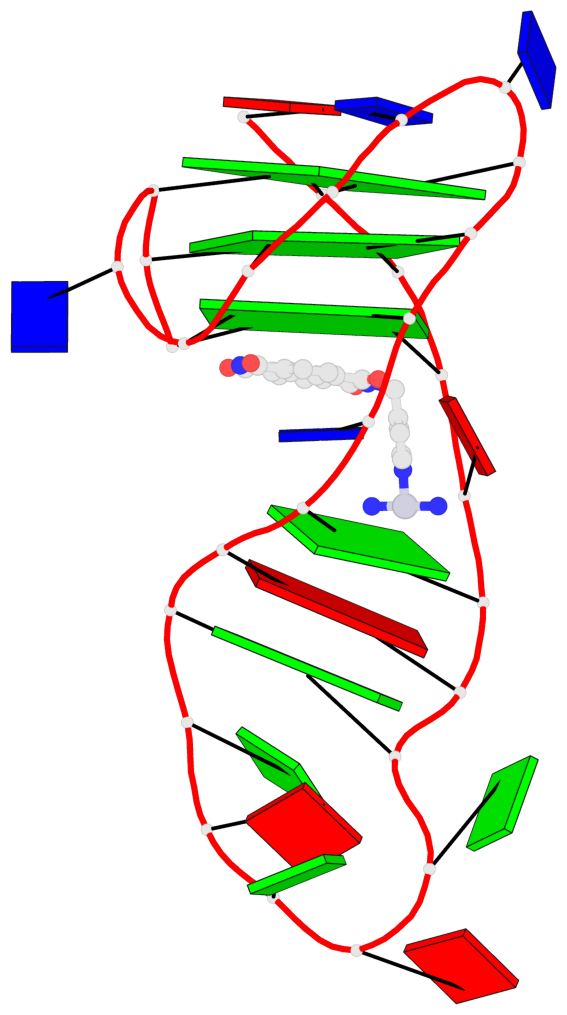
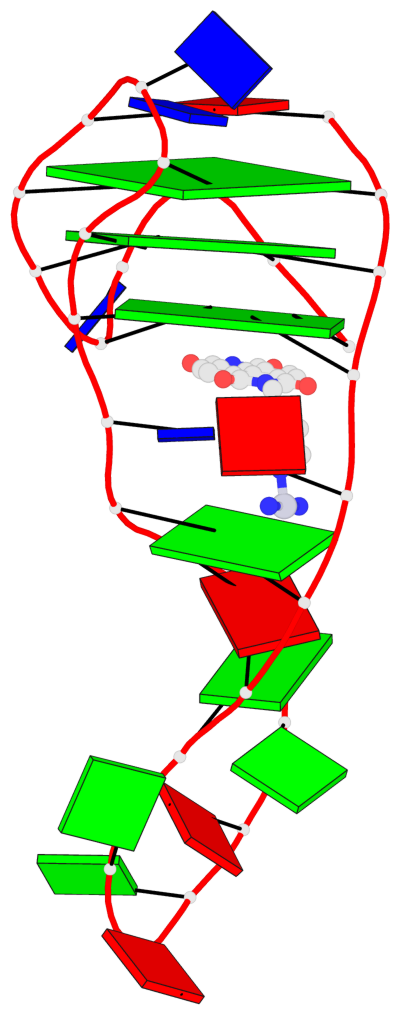
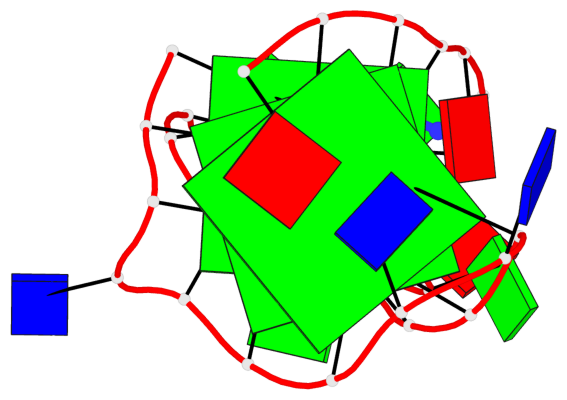
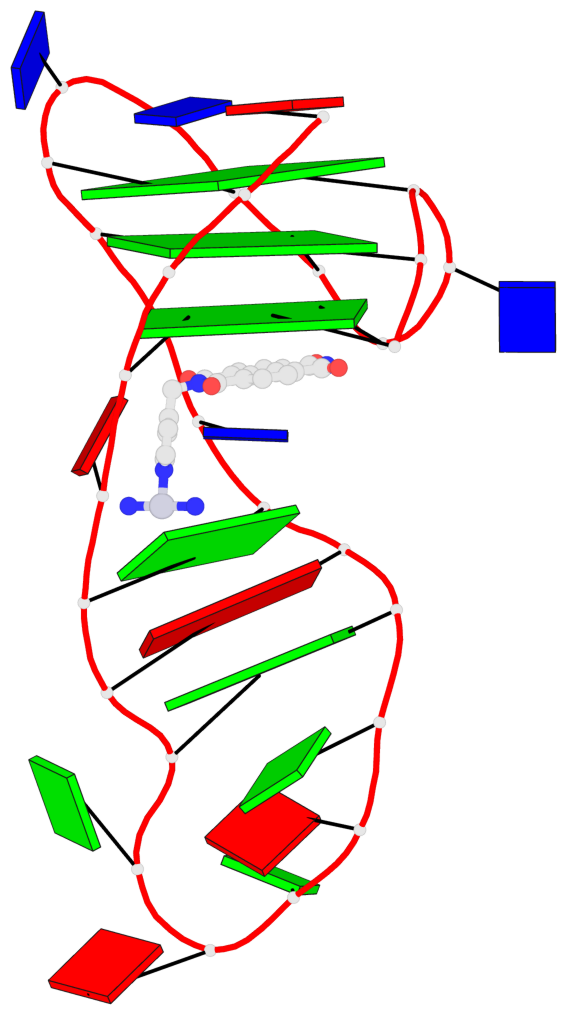
Detailed DSSR results for the G-quadruplex: PDB entry 8ijc
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8ijc
- Class
- DNA
- Method
- NMR
- Summary
- NMR solution structure of the 1:1 complex of a platinum(ii) ligand l1-transpt covalently bound to a g-quadruplex myt1l
- Reference
- Liu LY, Ma TZ, Zeng YL, Liu W, Zhang H, Mao ZW (2023): "Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy." Angew.Chem.Int.Ed.Engl., 62, e202305645. doi: 10.1002/anie.202305645.
- Abstract
- G-quadruplexes (G4s) have been revived as promising therapeutic targets with the development of immunotherapy, but the G4-mediated immune response remains unclear. We designed a novel class of G4-binding organic-platinum hybrids, L1 -cispt and L1 -transpt, with spatial matching for G4 binding and G4 DNA reactivity for binding site locking. The solution structure of L1 -transpt-MYT1L G4 demonstrated the effectiveness of the covalent binding and revealed the covalent binding-guided dynamic balance, accompanied by the destruction of the A5-T17 base pairs to achieve the covalent binding of the platinum unit to N7 of the G6 residue. Furthermore, L1 -cispt- and L1 -transpt-mediated genomic dysfunction could activate the retinoic acid-induced gene I (RIG-I) pathway and induce immunogenic cell death (ICD). The use of L1 -cispt/L1 -transpt-treated dying cells as therapeutic vaccines stimulated a robust immune response and effectively inhibited tumor growth in vivo. Our findings highlight the importance of the rational combination of specific spatial recognition and covalent locking in G4-trageting drug design and their potential in immunotherapy.
- G4 notes
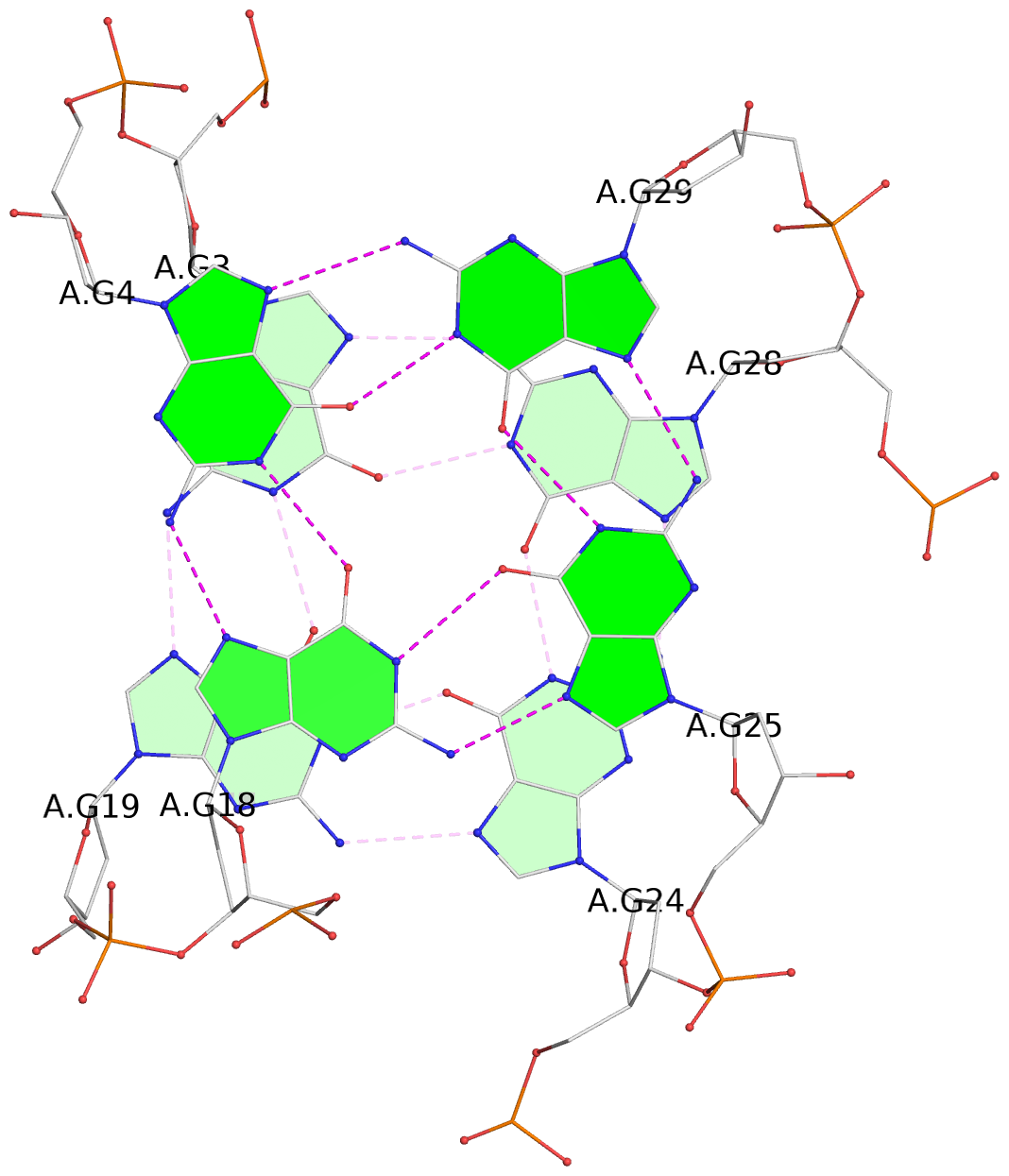
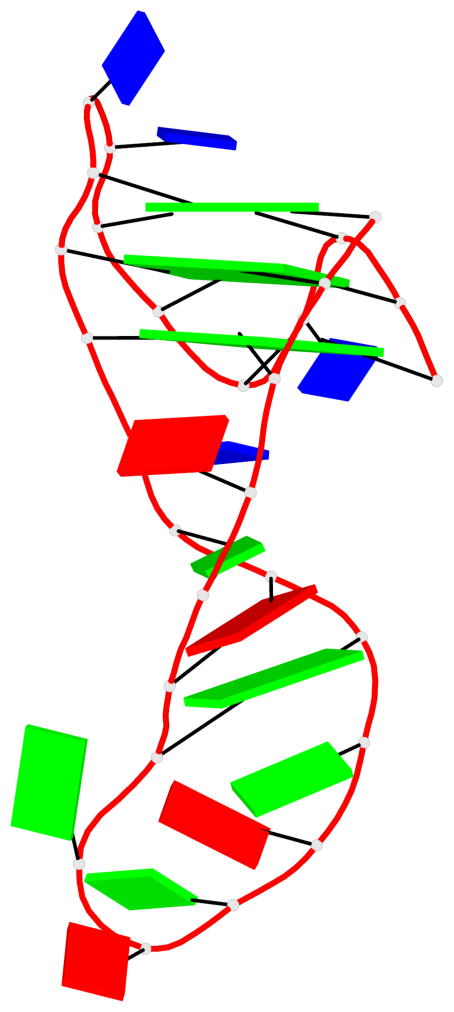
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
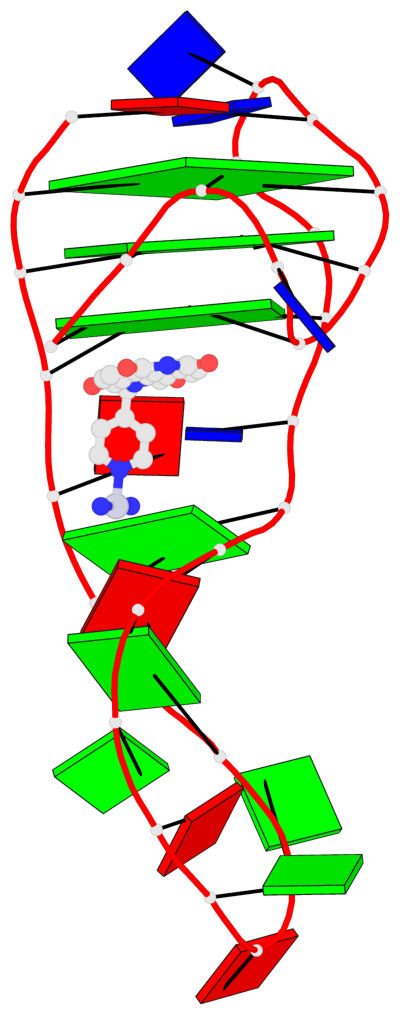
Base-block schematics in six views
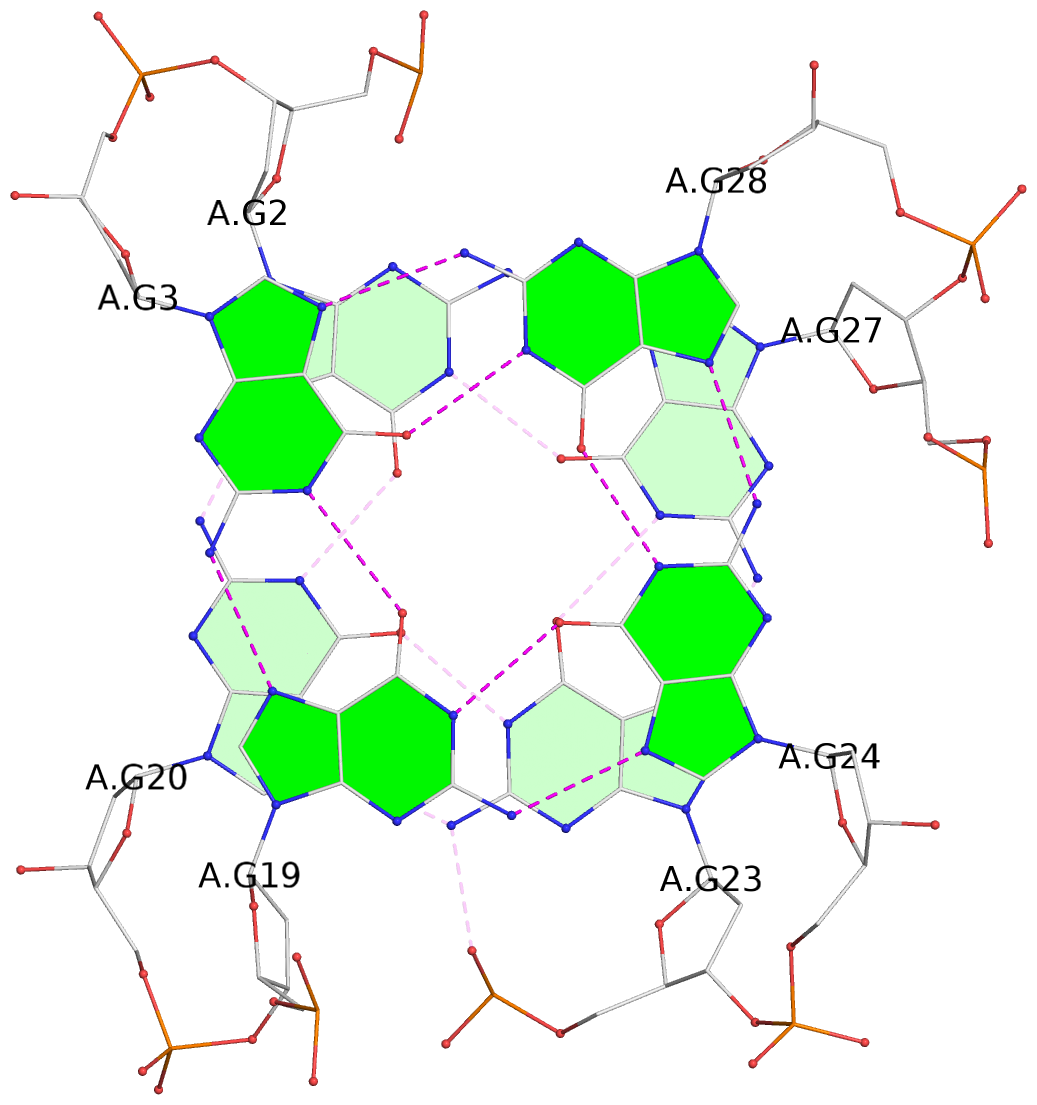
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=-... groove=wn-- planarity=0.245 type=bowl nts=4 GGGG A.DG2,A.DG20,A.DG23,A.DG27 2 glyco-bond=-s-- sugar=.... groove=wn-- planarity=0.193 type=other nts=4 GGGG A.DG3,A.DG19,A.DG24,A.DG28 3 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.294 type=bowl nts=4 GGGG A.DG4,A.DG18,A.DG25,A.DG29
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.