Detailed DSSR results for the G-quadruplex: PDB entry 8jfq
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8jfq
- Class
- DNA
- Method
- NMR
- Summary
- Structure of the major g-quadruplex in the human egfr oncogene promoter adopts a unique folding topology with a distinctive snap-back loop
- Reference
- Liu Y, Li J, Zhang Y, Wang Y, Chen J, Bian Y, Xia Y, Yang MH, Zheng K, Wang KB, Kong LY (2023): "Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop." J.Am.Chem.Soc., 145, 16228-16237. doi: 10.1021/jacs.3c05214.
- Abstract
- EGFR tyrosine kinase inhibitors have made remarkable success in targeted cancer therapy. However, therapeutic resistance inevitably occurred and EGFR-targeting therapy has been demonstrated to have limited efficacy or utility in glioblastoma, colorectal cancer, and hepatocellular carcinoma. Therefore, there is a high demand for the development of new targets to inhibit EGFR signaling. Herein, we found that the EGFR oncogene proximal promoter sequence forms a unique type of snap-back loop containing G-quadruplex (G4), which can be targeted by small molecules. For the first time, we determined the NMR solution structure of this snap-back EGFR-G4, a three-tetrad-core, parallel-stranded G4 with naturally occurring flanking residues at both the 5'-end and 3'-end. The snap-back loop located at the 3'-end region forms a stable capping structure through two stacked G-triads connected by multiple potential hydrogen bonds. Notably, the flanking residues are consistently absent in reported snap-back G4s, raising the question of whether such structures truly exist under in vivo conditions. The resolved EGFR-G4 structure has eliminated the doubt and showed distinct structural features that distinguish it from the previously reported snap-back G4s, which lack the flanking residues. Furthermore, we found that the snap-back EGFR-G4 structure is highly stable and can form on an elongated DNA template to inhibit DNA polymerase. The unprecedented high-resolution EGFR-G4 structure has thus contributed a promising molecular target for developing alternative EGFR signaling inhibitors in cancer therapeutics. Meanwhile, the two stacked triads may provide an attractive site for specific small-molecule targeting.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
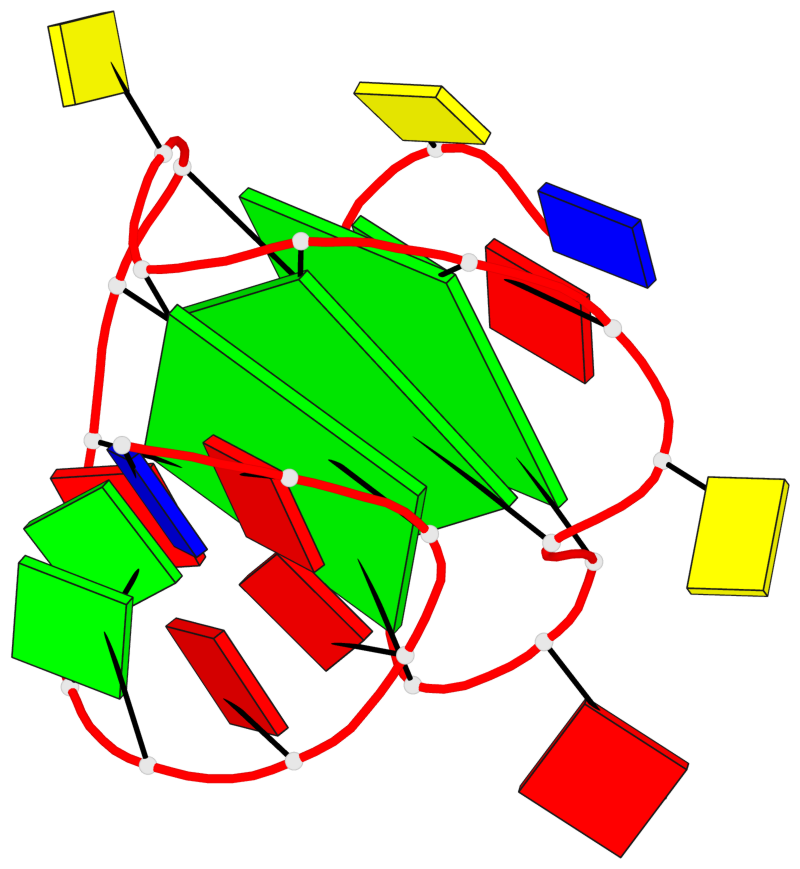
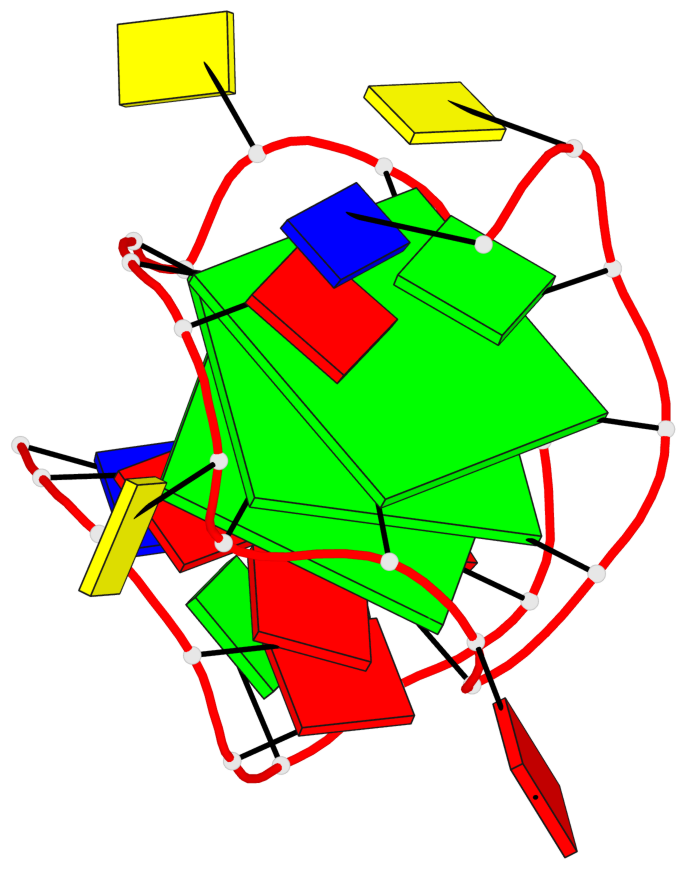
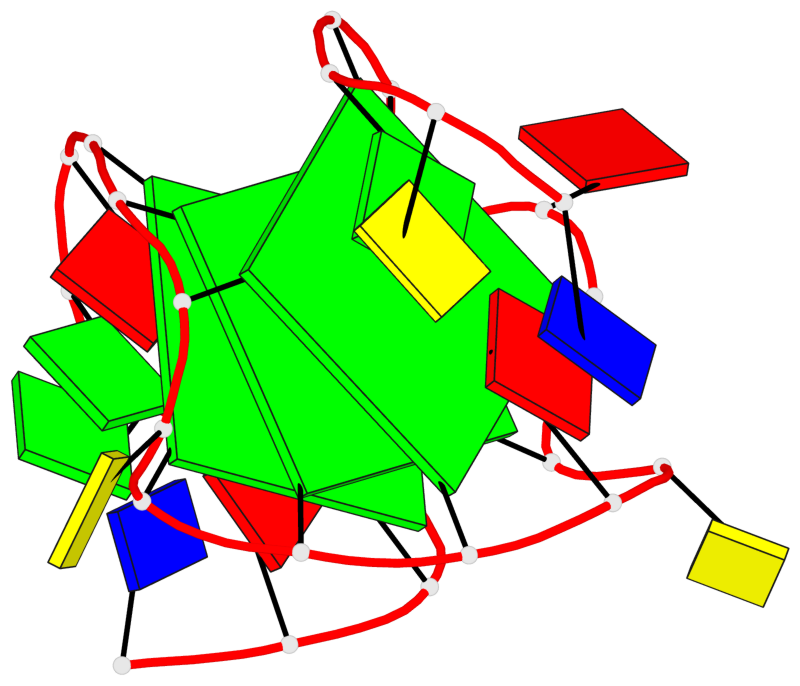
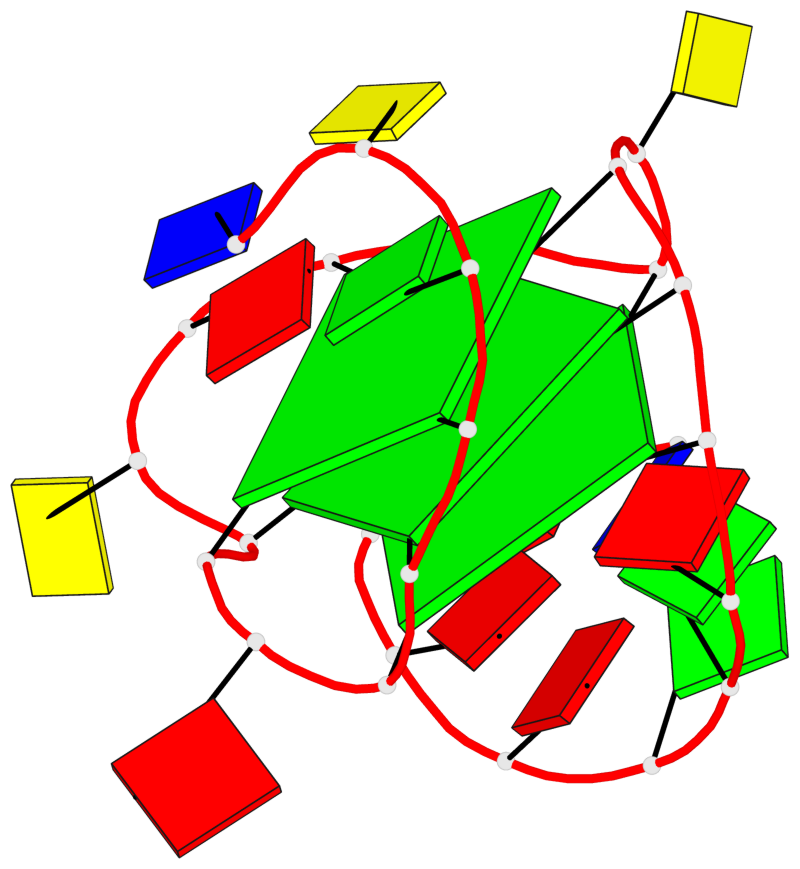
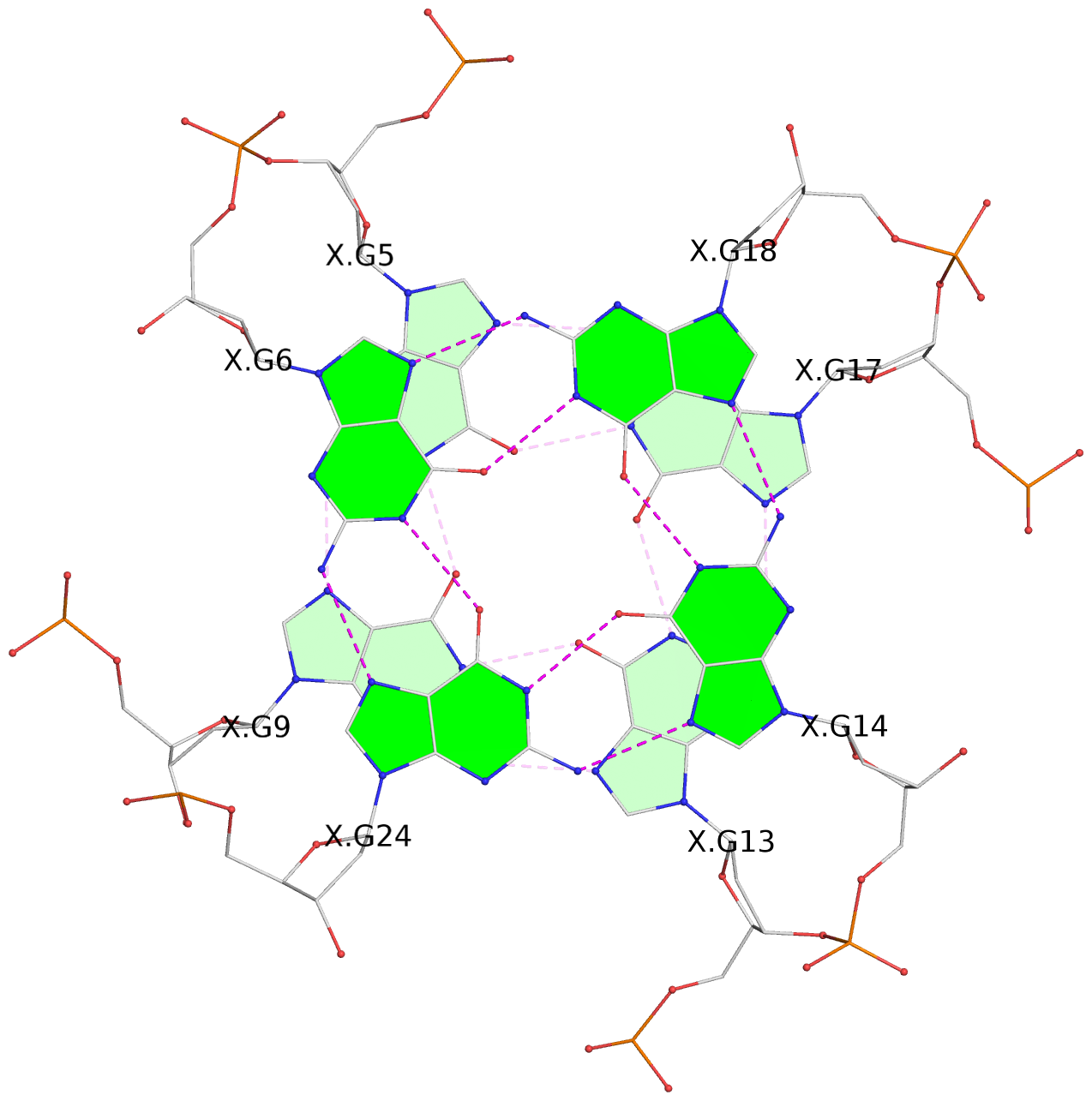
Base-block schematics in six views
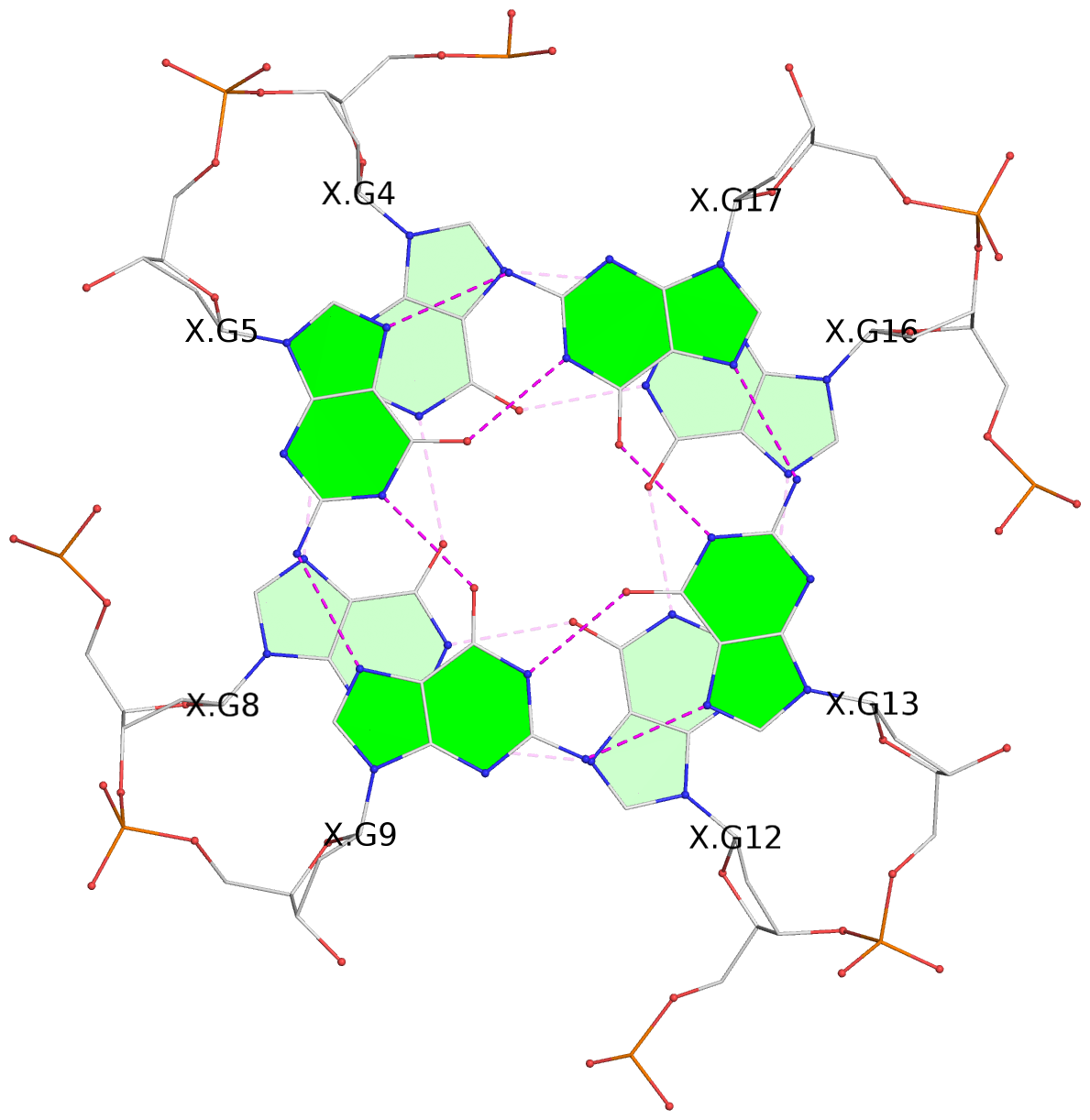
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.216 type=other nts=4 GGGG X.DG4,X.DG8,X.DG12,X.DG16 2 glyco-bond=---- sugar=---- groove=---- planarity=0.228 type=other nts=4 GGGG X.DG5,X.DG9,X.DG13,X.DG17 3 glyco-bond=---- sugar=---. groove=---- planarity=0.135 type=planar nts=4 GGGG X.DG6,X.DG24,X.DG14,X.DG18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 non-stem G4-loop (including the two closing Gs)
1 type=diagonal helix=#1 nts=7 GAGGAAG X.DG18,X.DA19,X.DG20,X.DG21,X.DA22,X.DA23,X.DG24