Detailed DSSR results for the G-quadruplex: PDB entry 8q4o
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8q4o
- Class
- RNA
- Method
- NMR
- Summary
- RNA g-quadruplex from the 5'-utr of human tyrosine kinase 2 (tyk2)
- Reference
- Orehova M, Plavec J, Kocman V (2024): "High-Resolution Structure of RNA G-Quadruplex Containing Unique Structural Motifs Originating from the 5'-UTR of Human Tyrosine Kinase 2 (TYK2)." Acs Omega, 9, 7215-7229. doi: 10.1021/acsomega.3c09592.
- Abstract
- Tyrosine kinase 2 (TYK2) is a member of the JAK family of nonreceptor-associated tyrosine kinases together with highly homologous JAK1, JAK2, and JAK3 paralogues. Overexpression of TYK2 is associated with several inflammatory diseases, including severe complications during the COVID-19 infection. Since the downregulation of JAK paralogues could lead to serious health consequences or even death, it is critical to avoid it when designing drugs to suppress TYK2. To achieve the required specificity only for TYK2, researchers have recently selectively targeted TYK2 mRNA by developing antisense oligonucleotides. In this work, we expand the target space of TYK2 mRNA by showing that the mRNA adopts tetra-helical noncanonical structures called G-quadruplexes. We identified a
- G4 notes
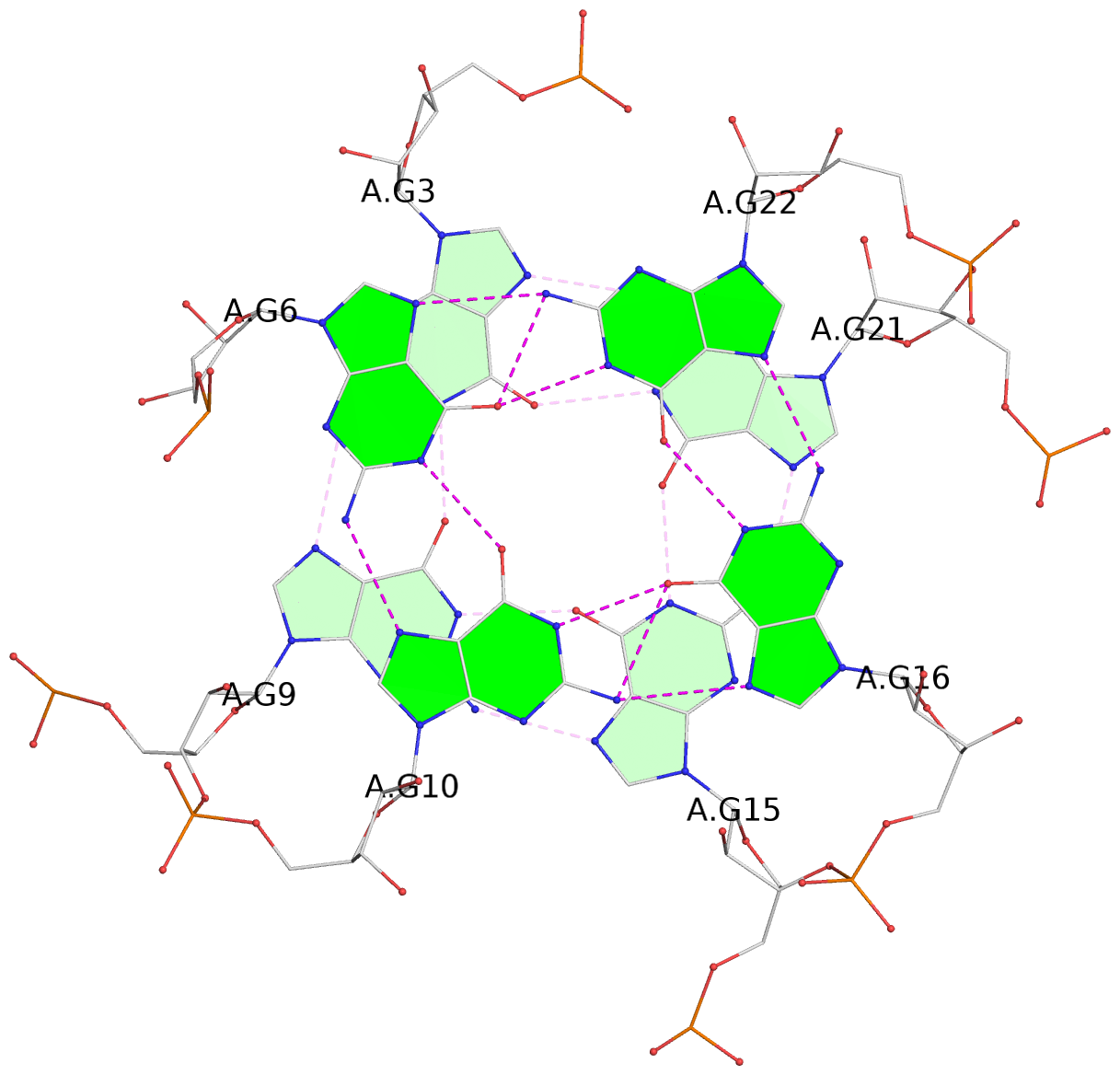
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
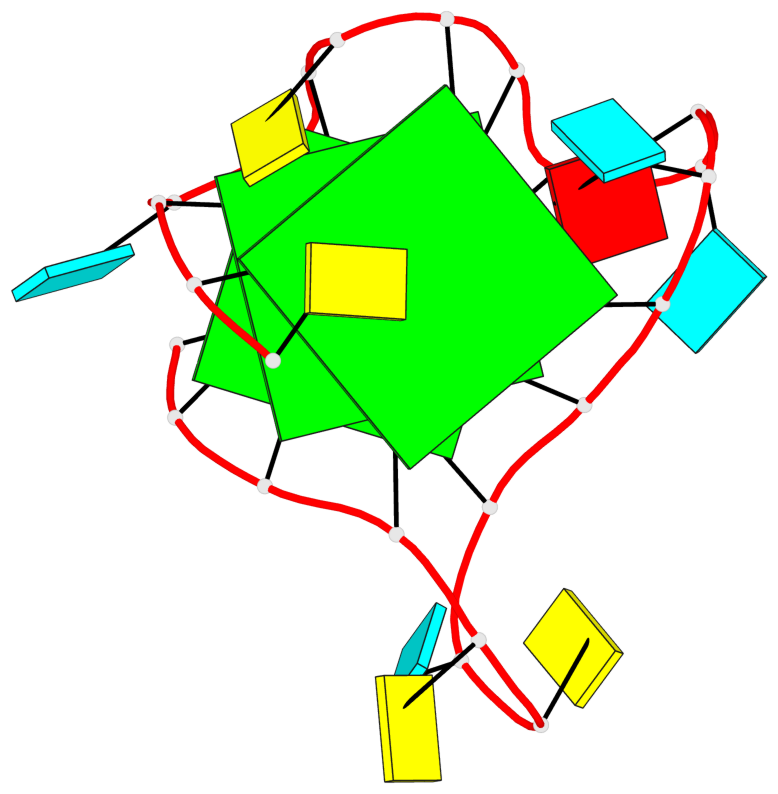
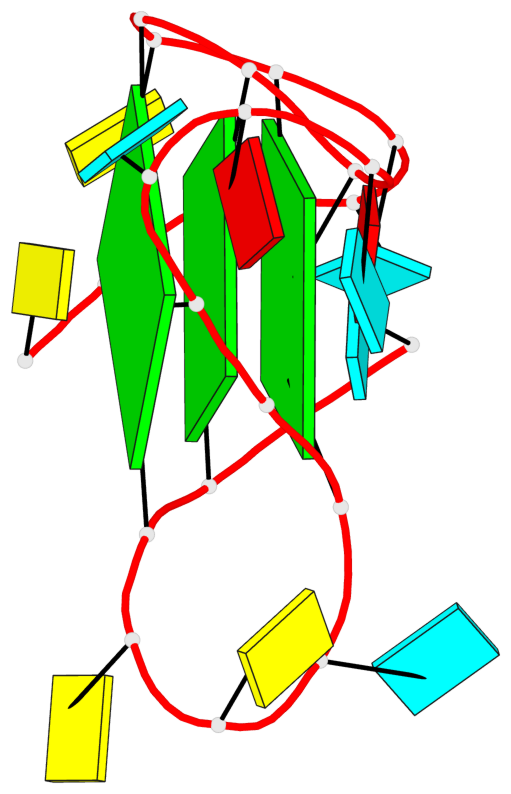
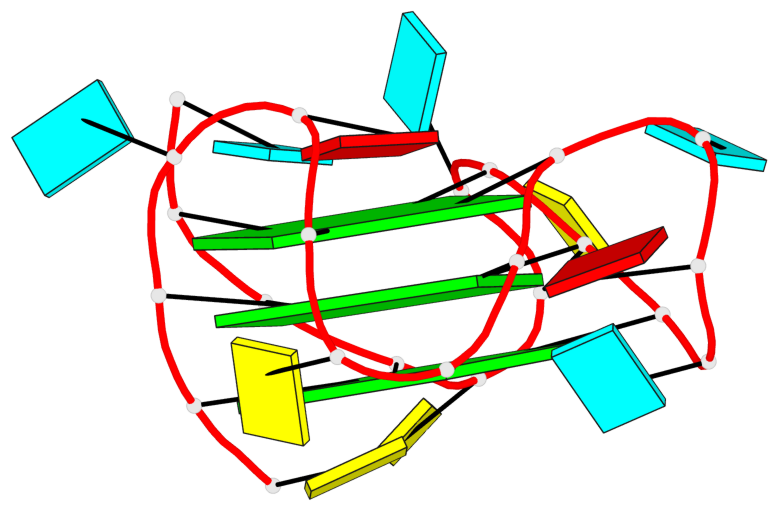
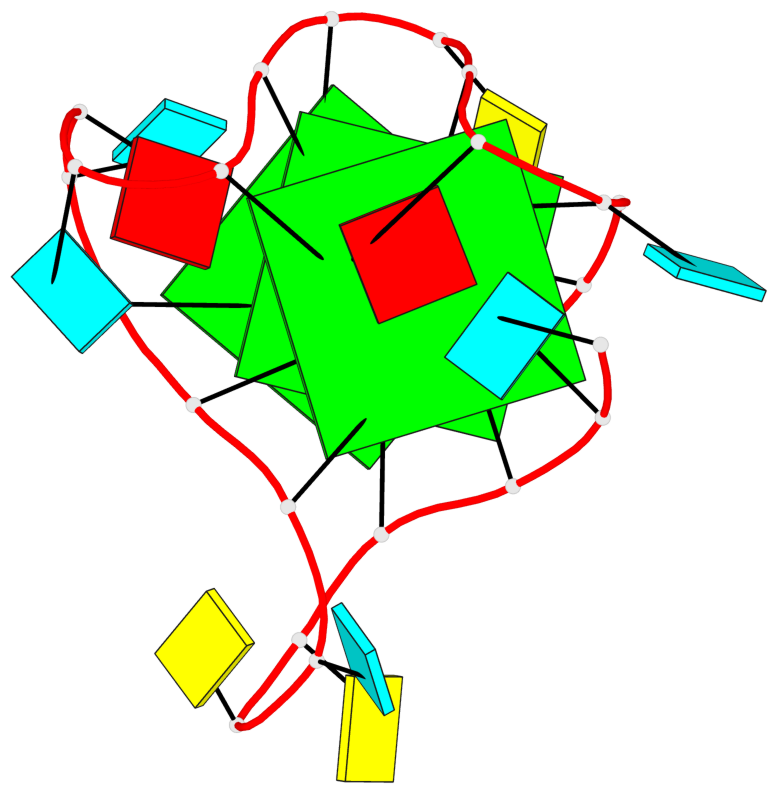
Base-block schematics in six views
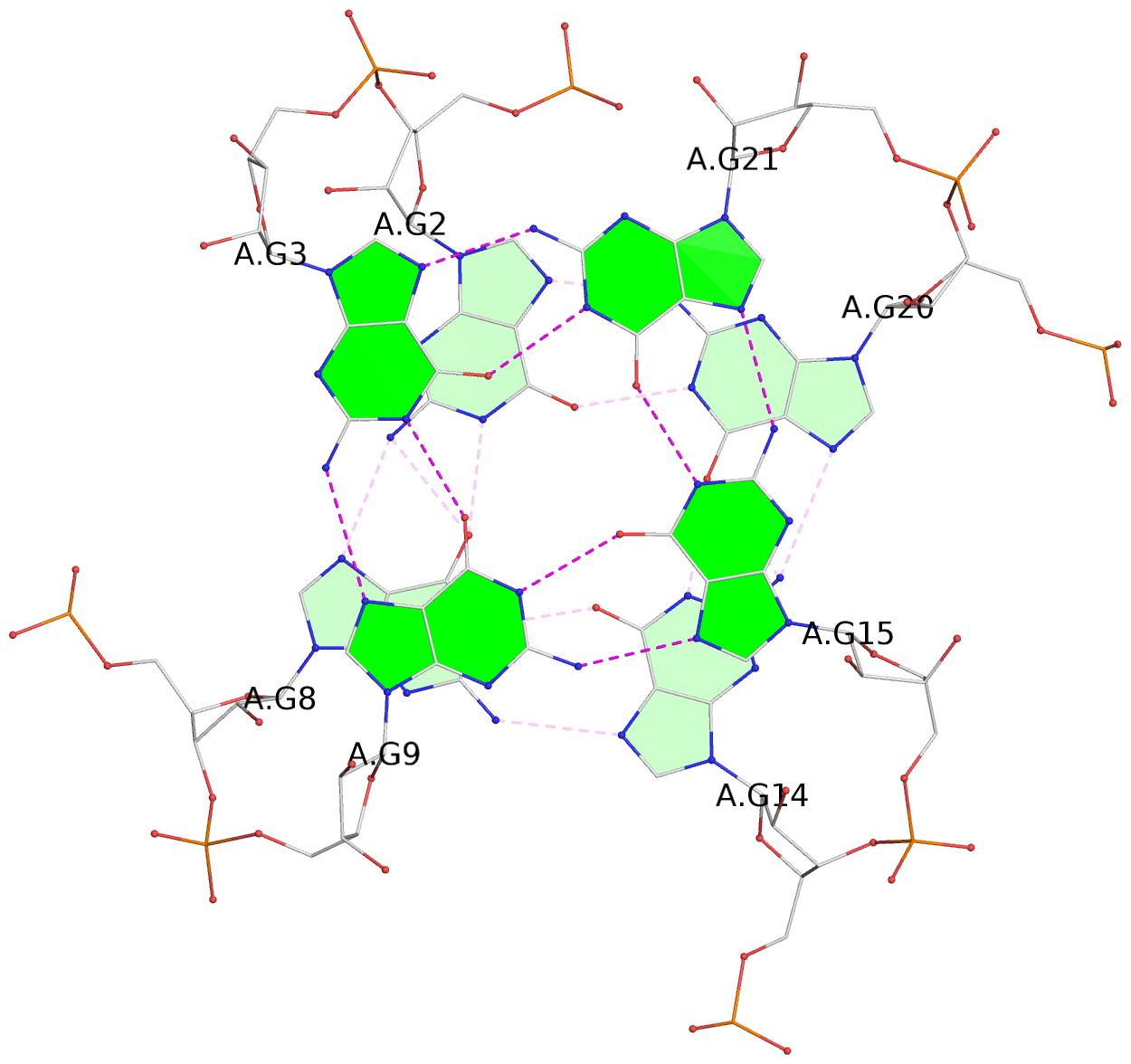
List of 3 G-tetrads
1 glyco-bond=---- sugar=3--- groove=---- planarity=0.146 type=planar nts=4 GGGG A.G2,A.G8,A.G14,A.G20 2 glyco-bond=---- sugar=3--3 groove=---- planarity=0.154 type=planar nts=4 GGGG A.G3,A.G9,A.G15,A.G21 3 glyco-bond=s--- sugar=33-- groove=w--n planarity=0.123 type=planar nts=4 GGGG A.G6,A.G22,A.G16,A.G10
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.