Detailed DSSR results for the G-quadruplex: PDB entry 8r4w
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8r4w
- Class
- DNA
- Method
- NMR
- Summary
- (3+1) hybrid-2 g-quadruplex with a -(llp) loop progression
- Reference
- Jana J, Vianney YM, Weisz K (2024): "Impact of loop length and duplex extensions on the design of hybrid-type G-quadruplexes." Chem.Commun.(Camb.), 60, 854-857. doi: 10.1039/d3cc05625b.
- Abstract
- A G-rich core sequence G3-TCA-G3-T1,2-G3-T1,2-G3 can be designed to fold into a parallel or into two different (3+1) hybrid-type G-quadruplexes, among them an elusive topology with one lateral followed by two propeller loops. Favored folds can be rationalized based on the number of intervening thymidines and on additional complementary flanking sequences.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
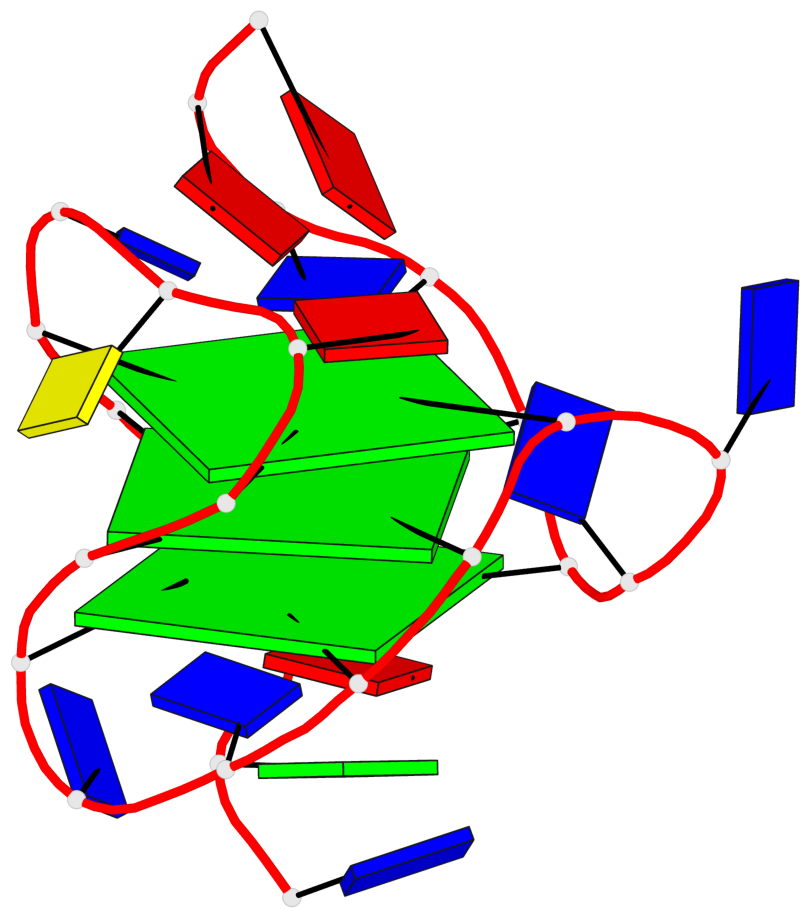
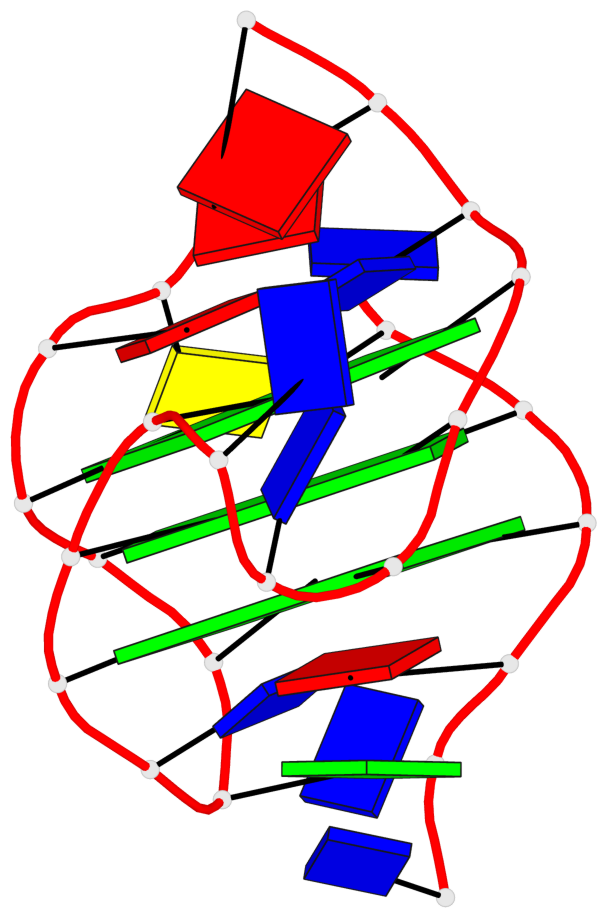
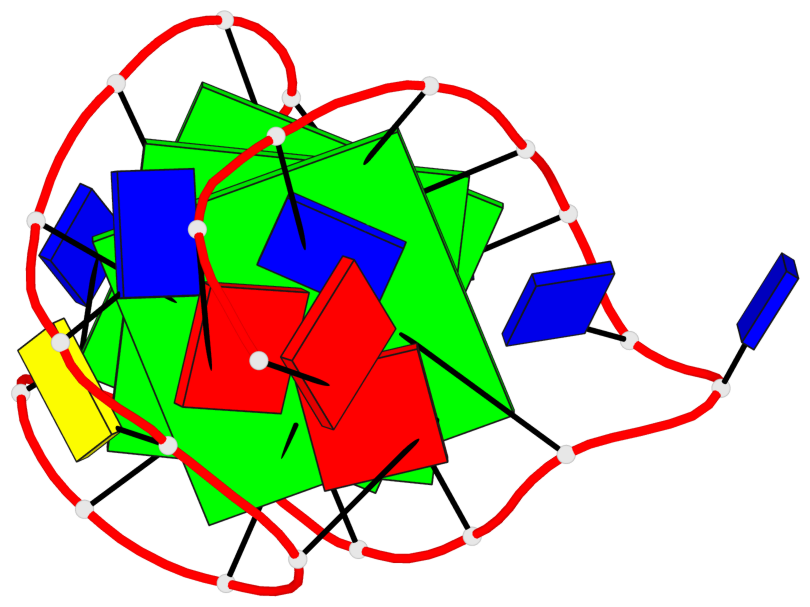
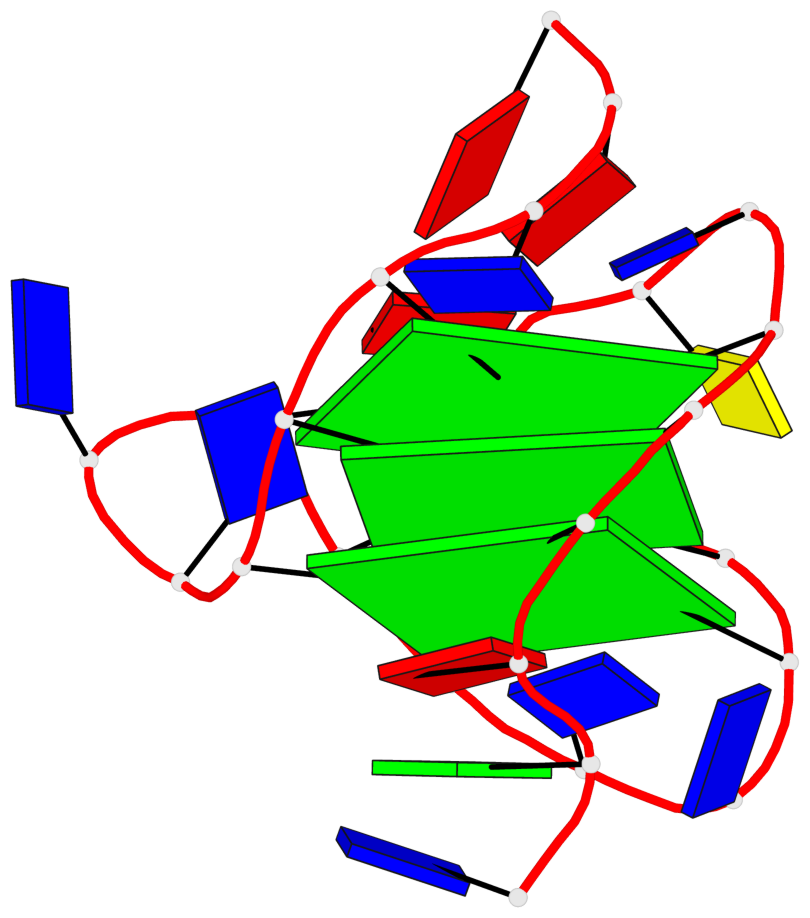
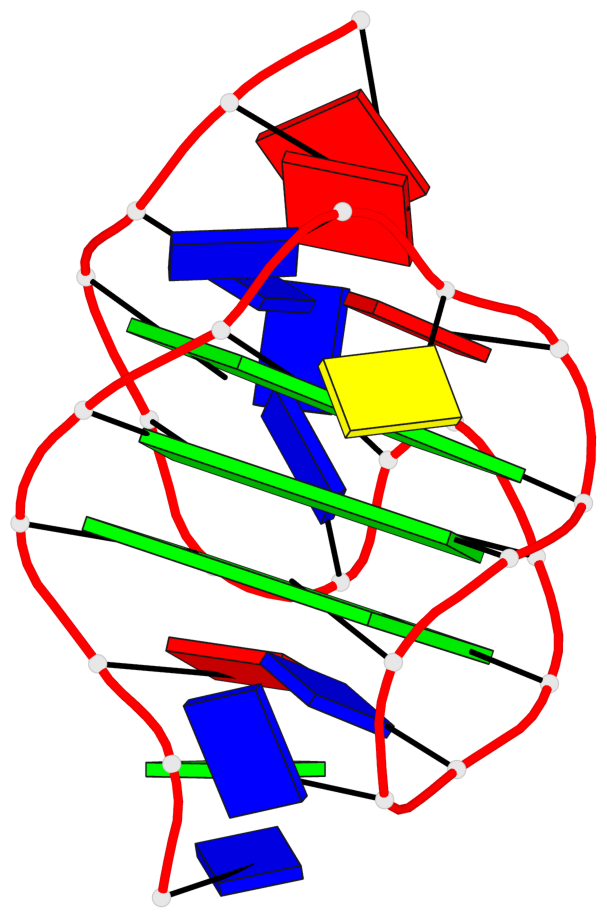
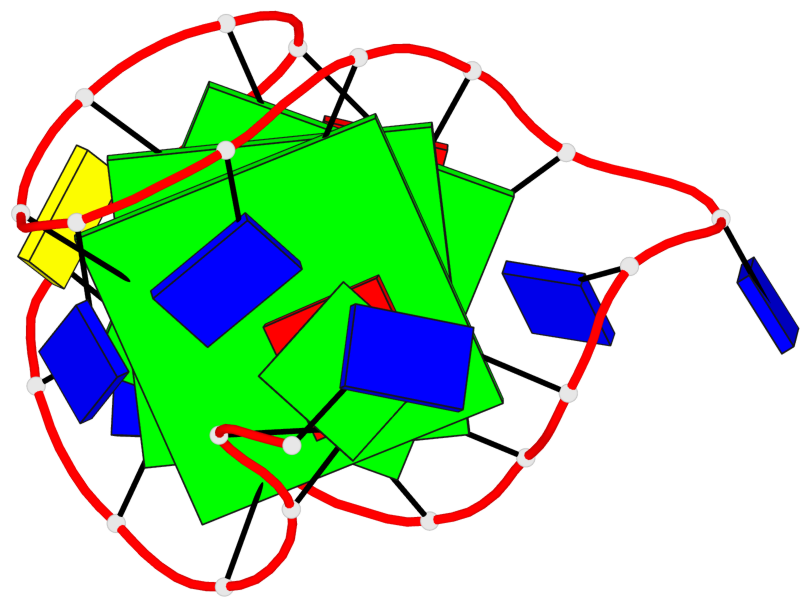
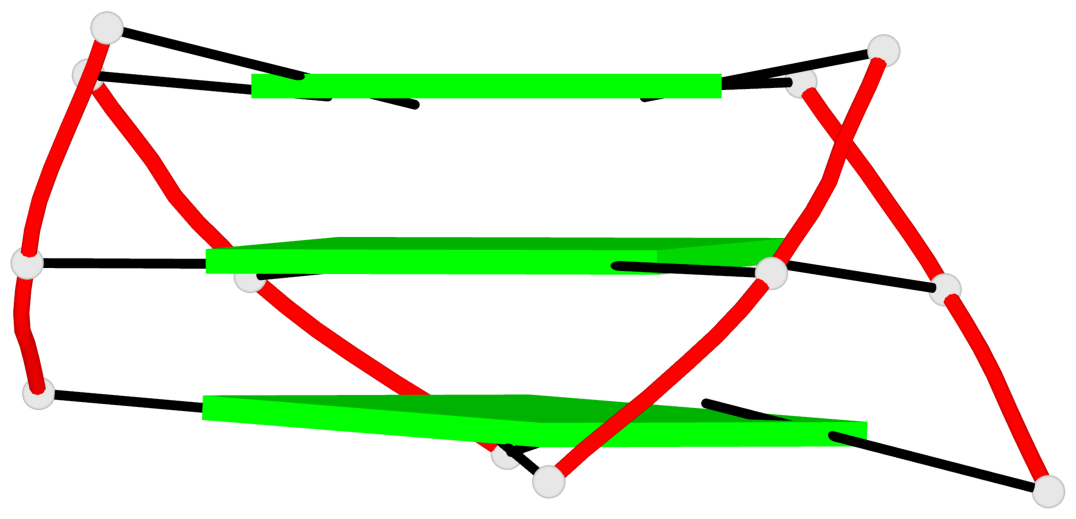
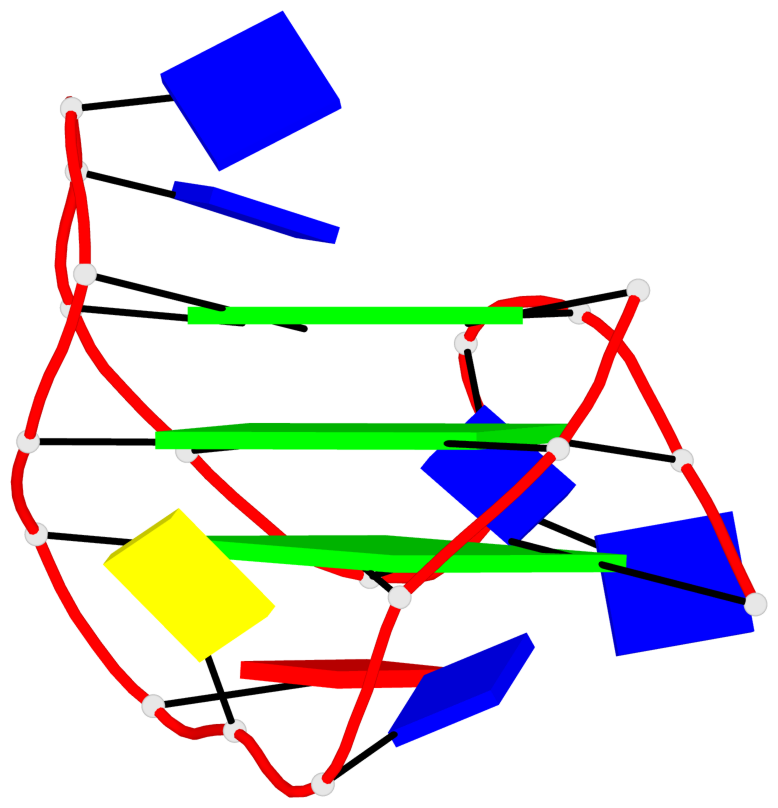
Base-block schematics in six views
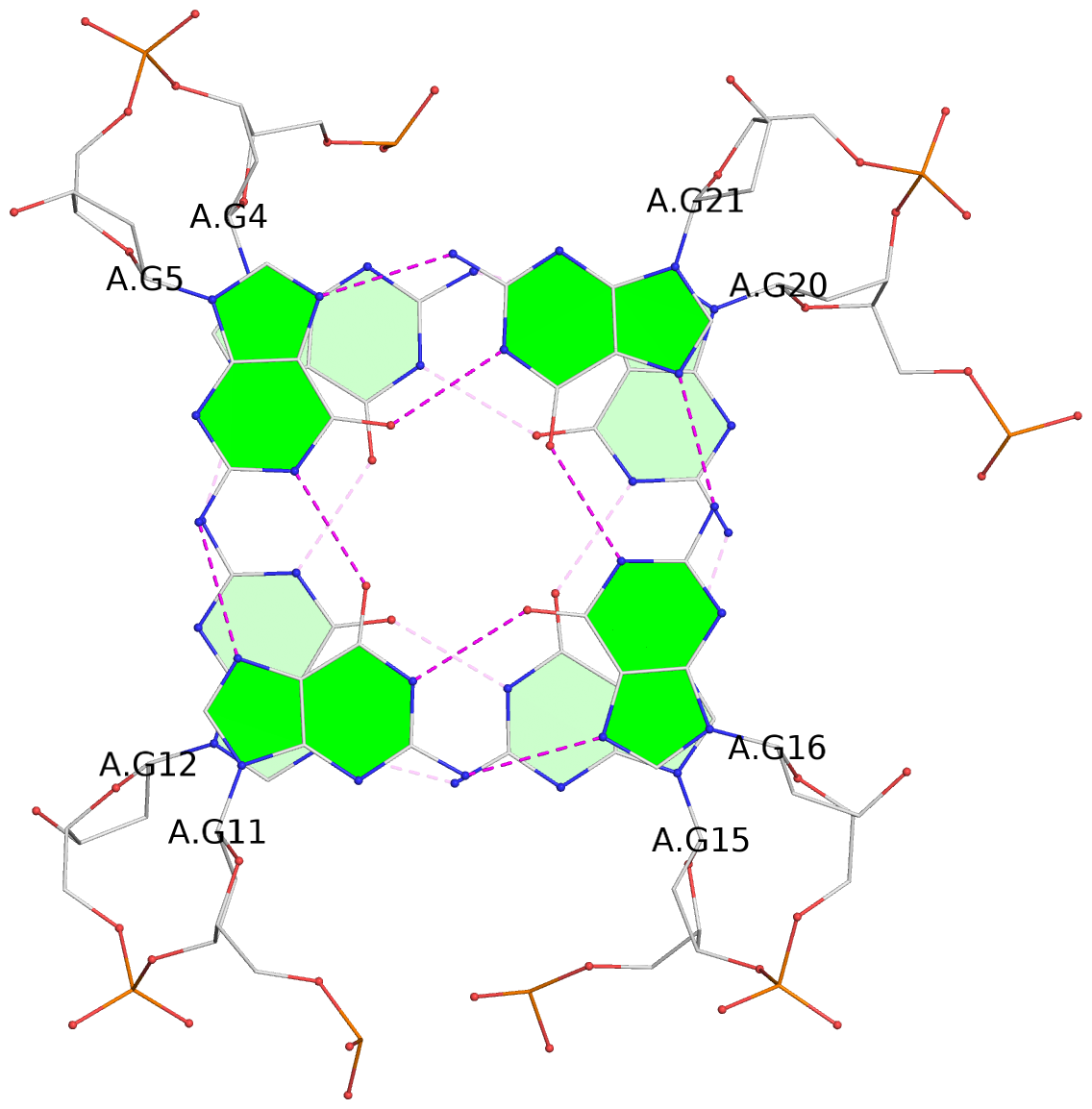
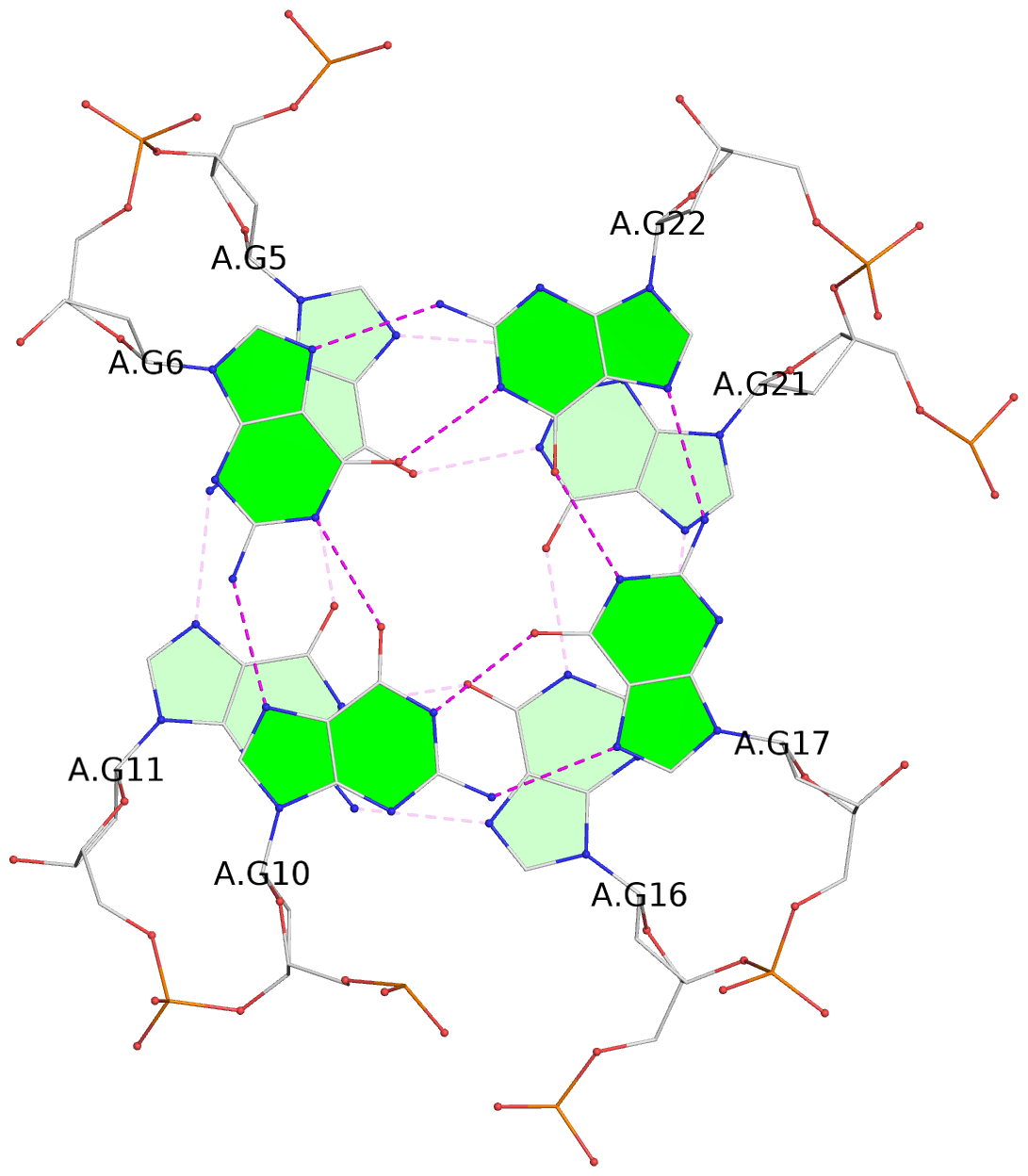
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=---- groove=wn-- planarity=0.228 type=other nts=4 GGGG A.DG4,A.DG12,A.DG15,A.DG20 2 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.139 type=planar nts=4 GGGG A.DG5,A.DG11,A.DG16,A.DG21 3 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.169 type=other nts=4 GGGG A.DG6,A.DG10,A.DG17,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.