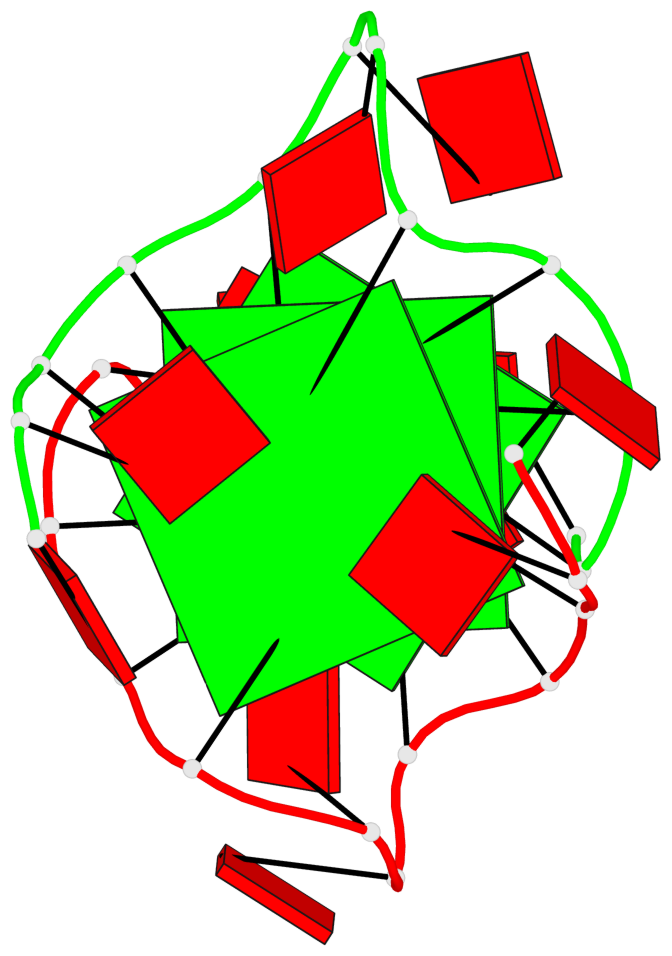
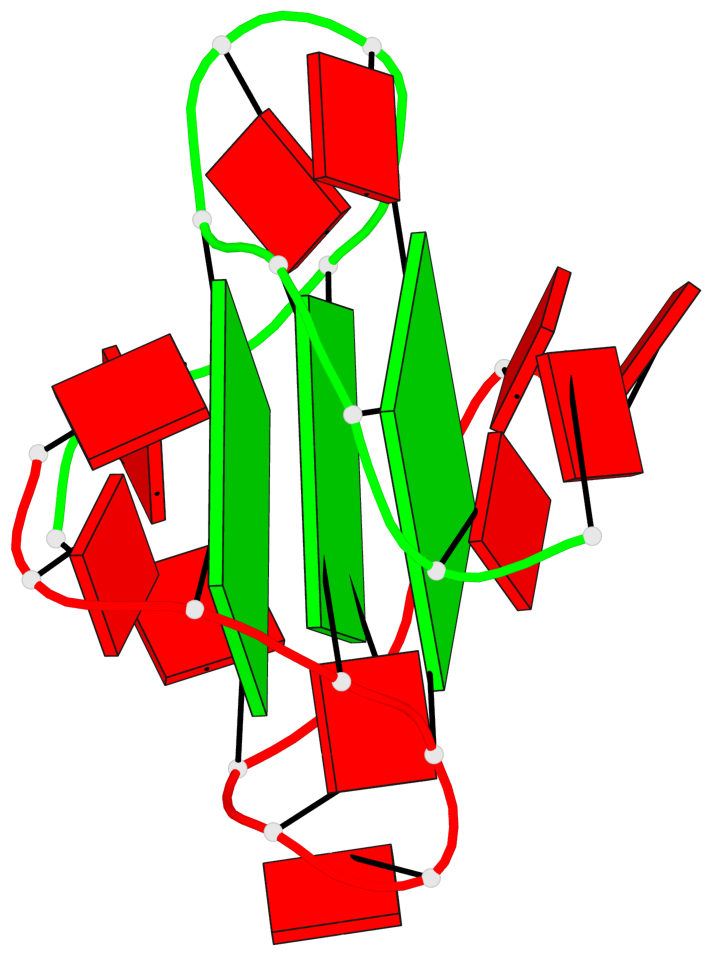
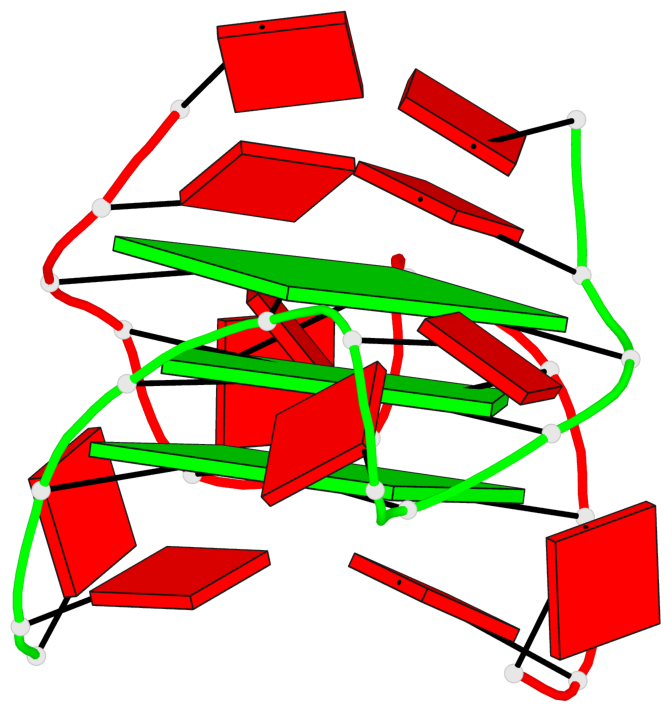
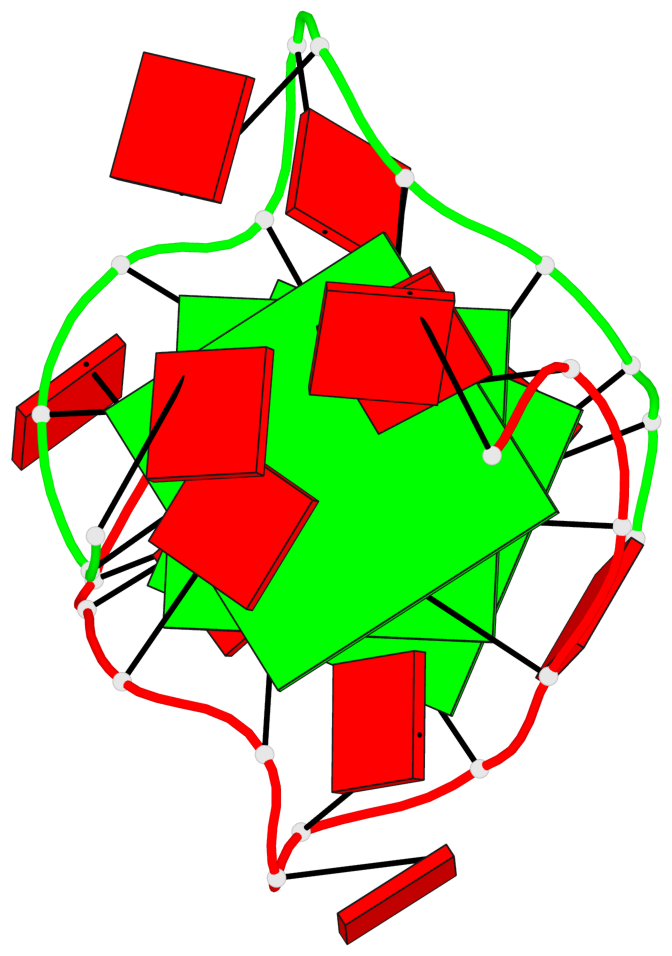
Detailed DSSR results for the G-quadruplex: PDB entry 8x1v
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8x1v
- Class
- DNA
- Method
- NMR
- Summary
- NMR structure of a bimolecular parallel g-quadruplex formed by aaggg repeats from pathogenic rfc1 gene
- Reference
- Wang Y, Wang J, Yan Z, Hou J, Wan L, Yang Y, Liu Y, Yi J, Guo P, Han D (2024): "Structural investigation of pathogenic RFC1 AAGGG pentanucleotide repeats reveals a role of G-quadruplex in dysregulated gene expression in CANVAS." Nucleic Acids Res., 52, 2698-2710. doi: 10.1093/nar/gkae032.
- Abstract
- An expansion of AAGGG pentanucleotide repeats in the replication factor C subunit 1 (RFC1) gene is the genetic cause of cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS), and it also links to several other neurodegenerative diseases including the Parkinson's disease. However, the pathogenic mechanism of RFC1 AAGGG repeat expansion remains enigmatic. Here, we report that the pathogenic RFC1 AAGGG repeats form DNA and RNA parallel G-quadruplex (G4) structures that play a role in impairing biological processes. We determine the first high-resolution nuclear magnetic resonance (NMR) structure of a bimolecular parallel G4 formed by d(AAGGG)2AA and reveal how AAGGG repeats fold into a higher-order structure composed of three G-tetrad layers, and further demonstrate the formation of intramolecular G4s in longer DNA and RNA repeats. The pathogenic AAGGG repeats, but not the nonpathogenic AAAAG repeats, form G4 structures to stall DNA replication and reduce gene expression via impairing the translation process in a repeat-length-dependent manner. Our results provide an unprecedented structural basis for understanding the pathogenic mechanism of AAGGG repeat expansion associated with CANVAS. In addition, the high-resolution structures resolved in this study will facilitate rational design of small-molecule ligands and helicases targeting G4s formed by AAGGG repeats for therapeutic interventions.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
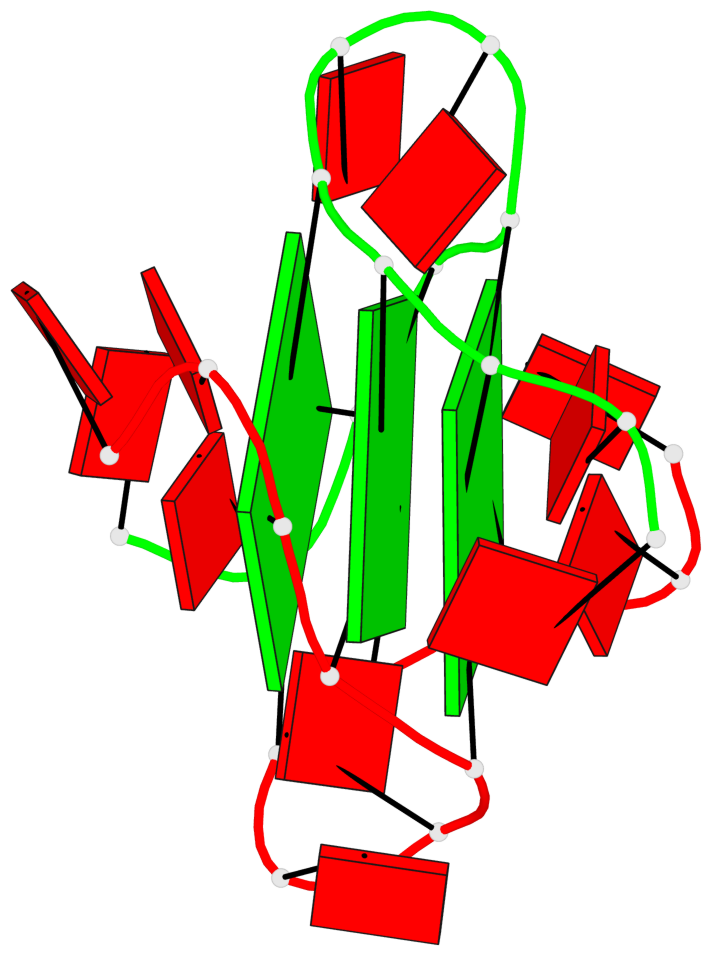
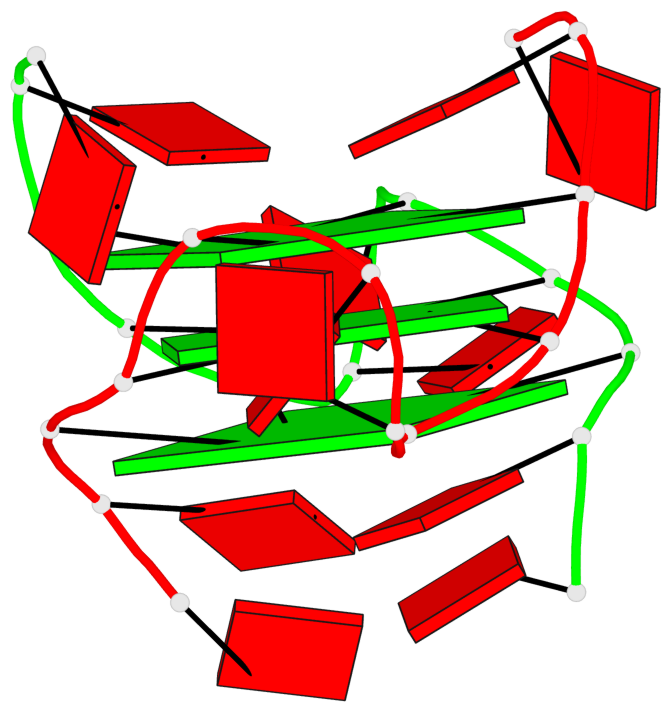
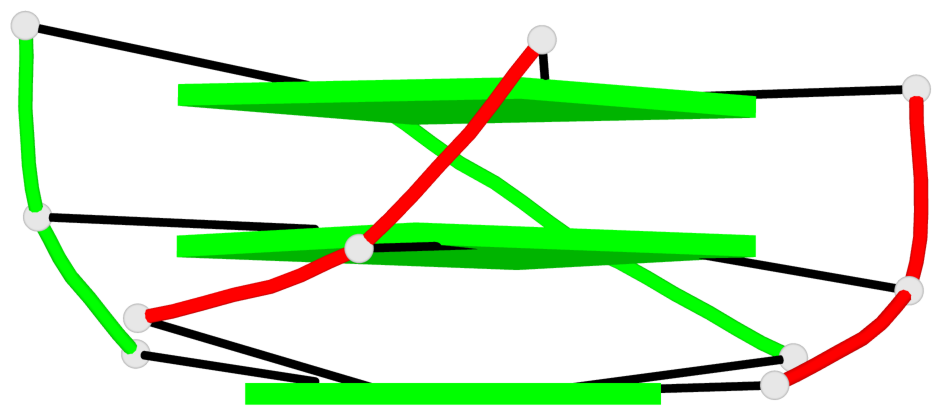
Base-block schematics in six views
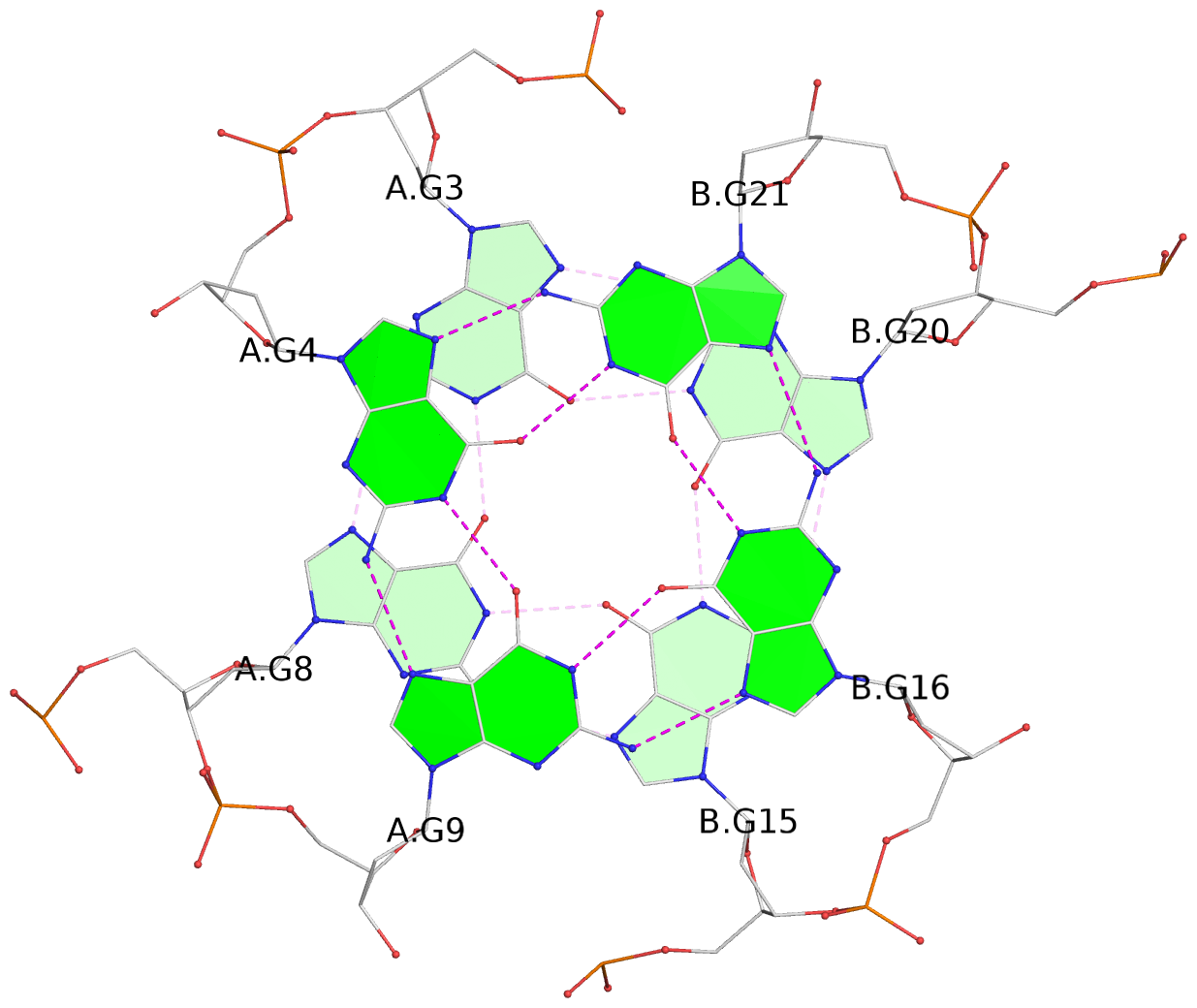
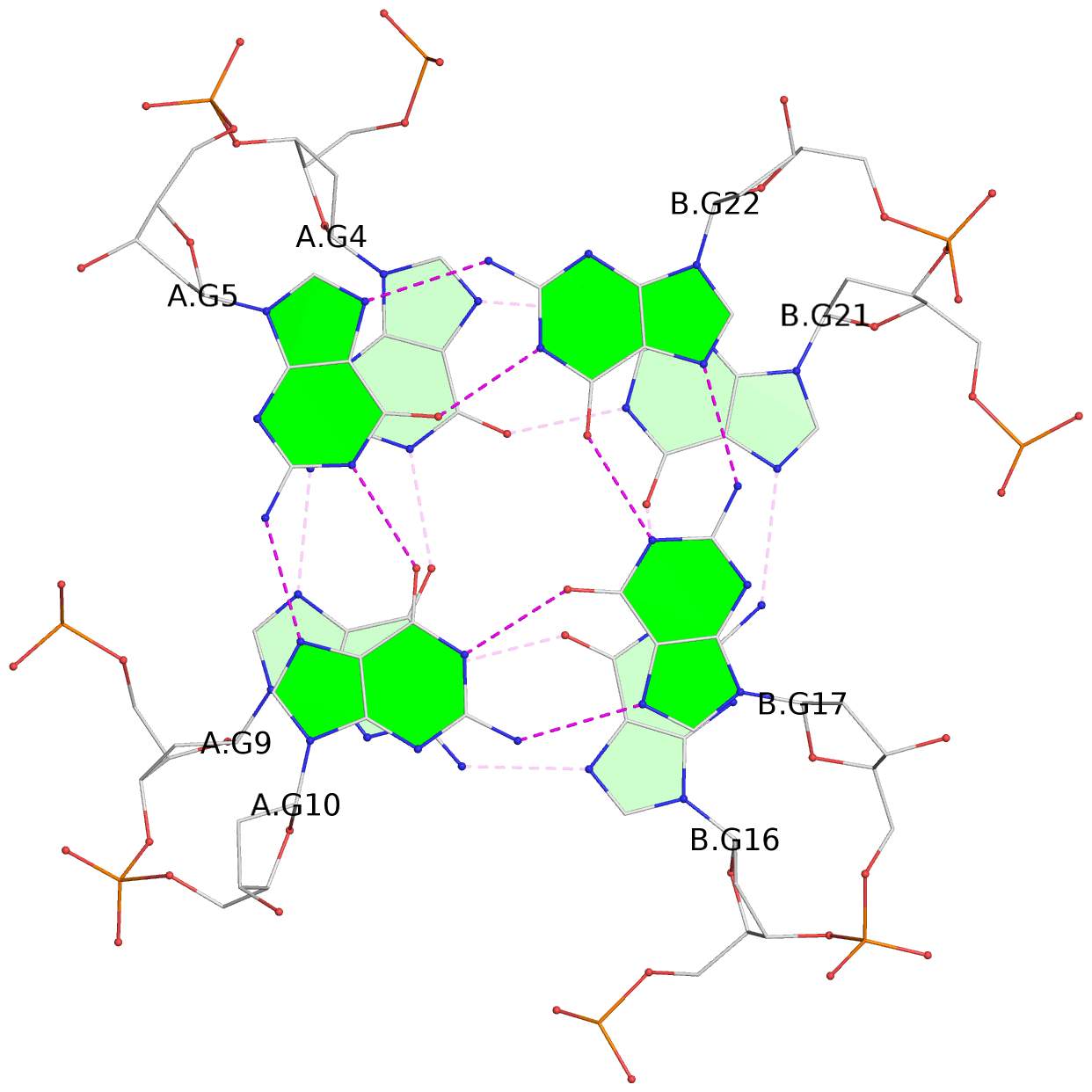
List of 3 G-tetrads
1 glyco-bond=---- sugar=---. groove=---- planarity=0.322 type=saddle nts=4 GGGG A.DG3,A.DG8,B.DG15,B.DG20 2 glyco-bond=---- sugar=---3 groove=---- planarity=0.215 type=other nts=4 GGGG A.DG4,A.DG9,B.DG16,B.DG21 3 glyco-bond=---- sugar=--.. groove=---- planarity=0.183 type=other nts=4 GGGG A.DG5,A.DG10,B.DG17,B.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.