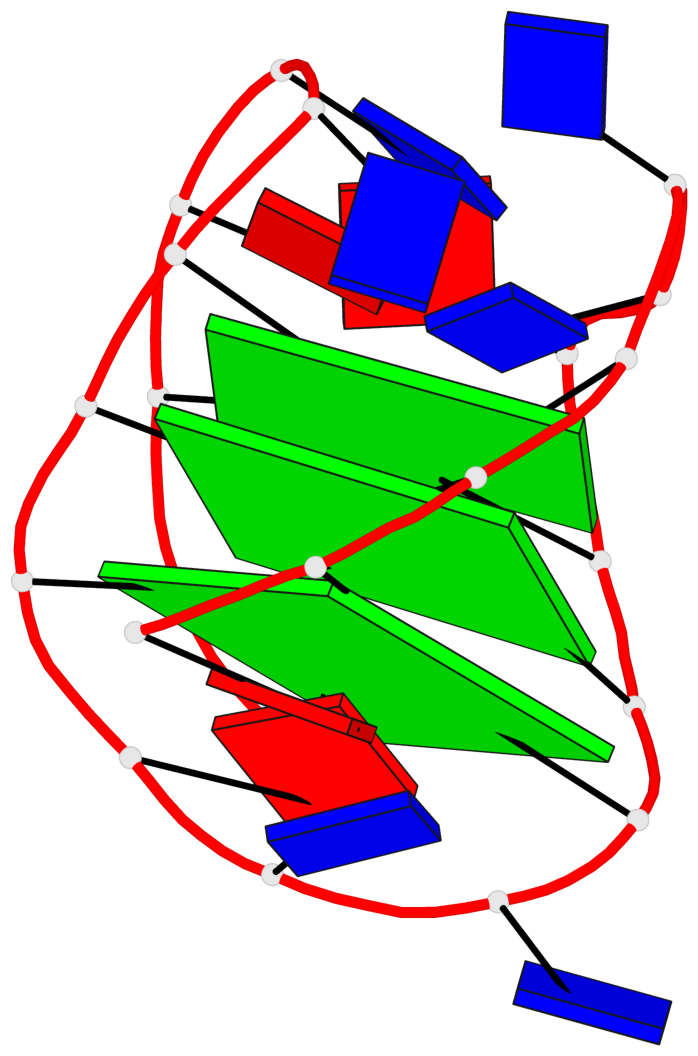
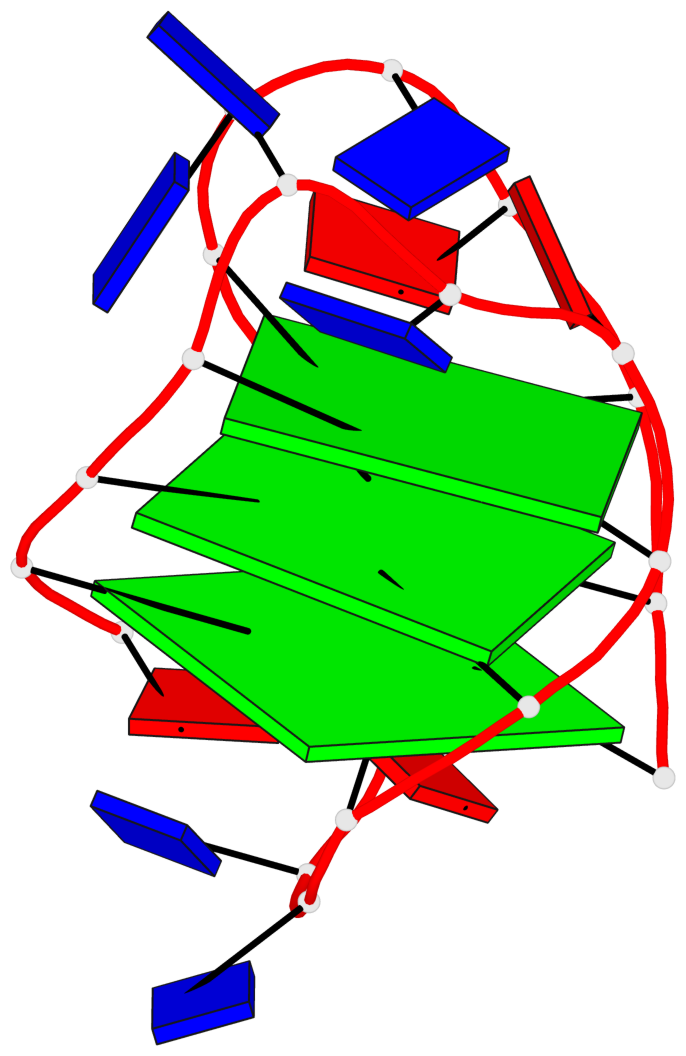
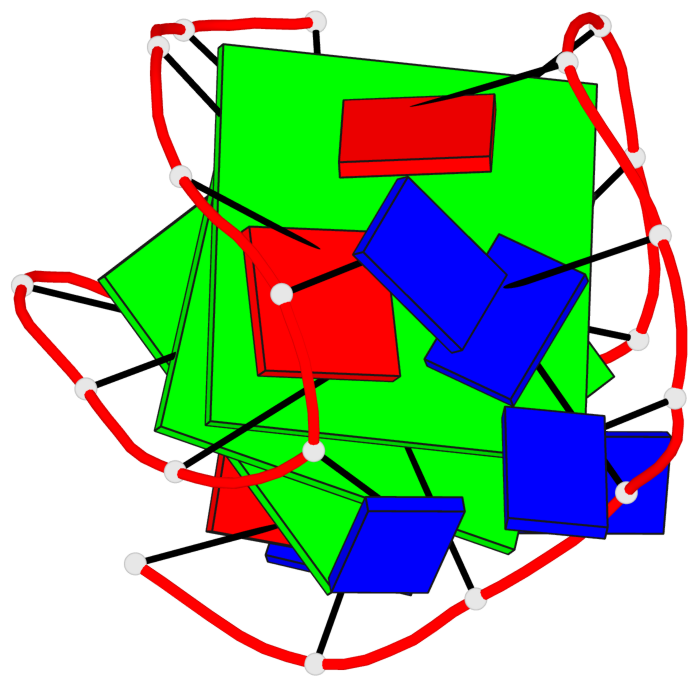
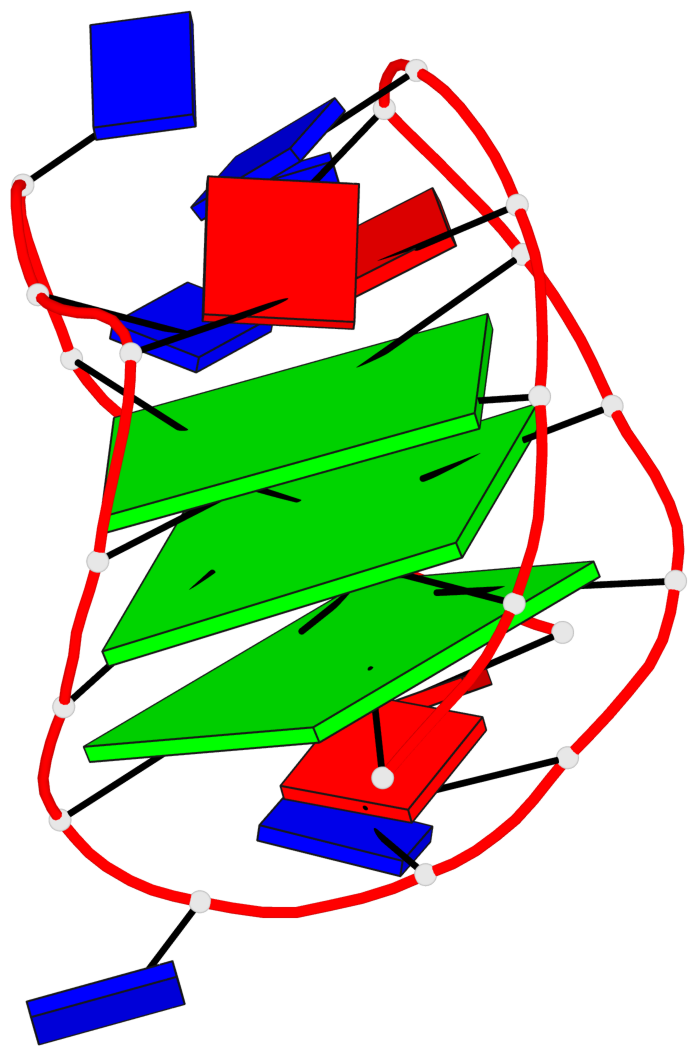
Detailed DSSR results for the G-quadruplex: PDB entry 143d
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 143d
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of the human telomeric repeat d(ag3[t2ag3]3) of the g-quadruplex
- Reference
- Wang Y, Patel DJ (1993): "Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex." Structure, 1, 263-282. doi: 10.1016/0969-2126(93)90015-9.
- Abstract
- Background: Repeats of Gn sequences are detected as single strand overhangs at the ends of eukaryotic chromosomes together with associated binding proteins. Such telomere sequences have been implicated in the replication and maintenance of chromosomal termini. They may also mediate chromosomal organization and association during meiosis and mitosis.
Results: We have determined the three-dimensional solution structure of the human telomere sequence, d[AG3(T2AG3)3] in Na(+)-containing solution using a combined NMR, distance geometry and molecular dynamics approach (including relaxation matrix refinement). The sequence, which contains four AG3 repeats, folds intramolecularly into a G-tetraplex stabilized by three stacked G-tetrads which are connected by two lateral loops and a central diagonal loop. Of the four grooves that are formed, one is wide, two are of medium width and one is narrow. The alignment of adjacent G-G-G segments in parallel generates the two grooves of medium width whilst the antiparallel arrangement results in one wide and one narrow groove. Three of the four adenines stack on top of adjacent G-tetrads while the majority of the thymines sample multiple conformations.
Conclusions: The availability of the d[AG3(T2AG3)3] solution structure containing four AG3 human telomeric repeats should permit the rational design of ligands that recognize and bind with specificity and affinity to the individual grooves of the G-tetraplex, as well as to either end containing the diagonal and lateral loops. Such ligands could modulate the equilibrium between folded G-tetraplex structures and their unfolded extended counterparts. - G4 notes
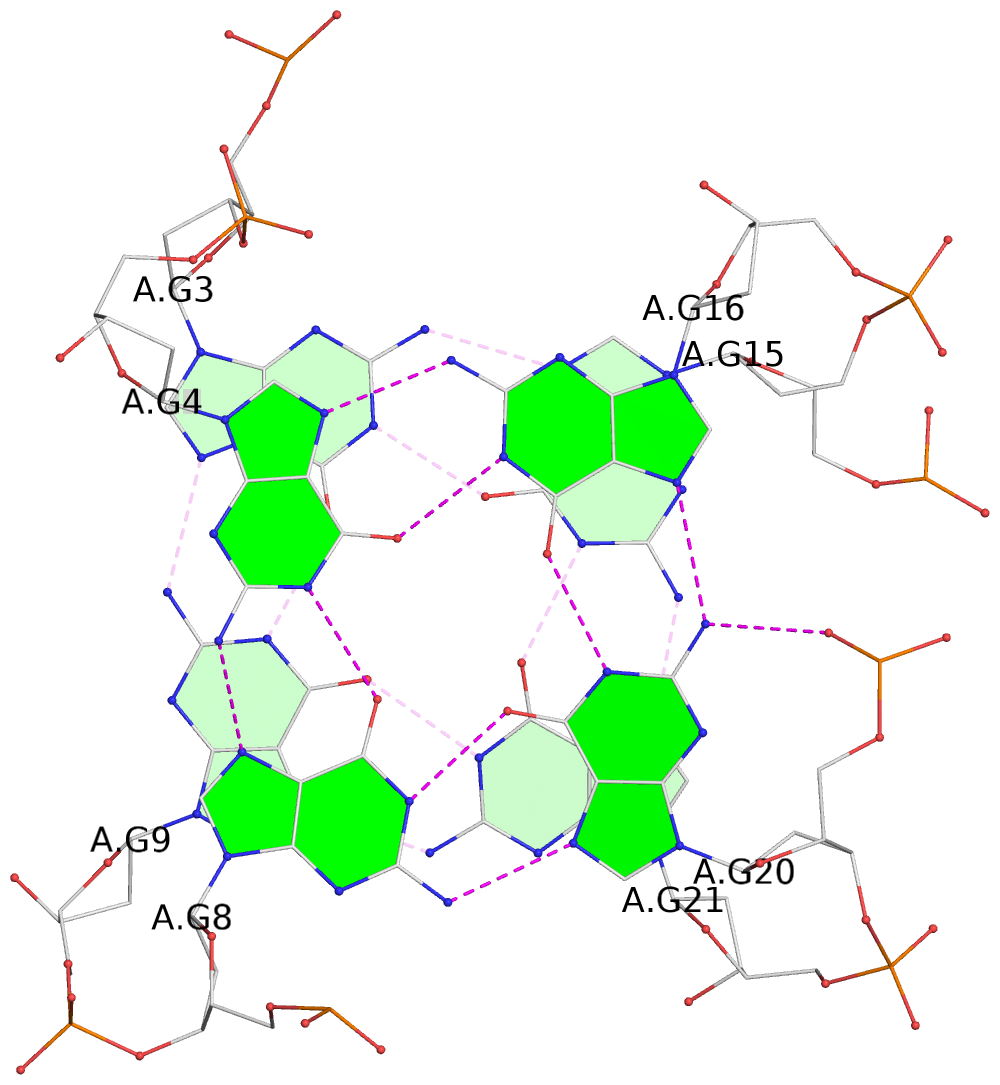
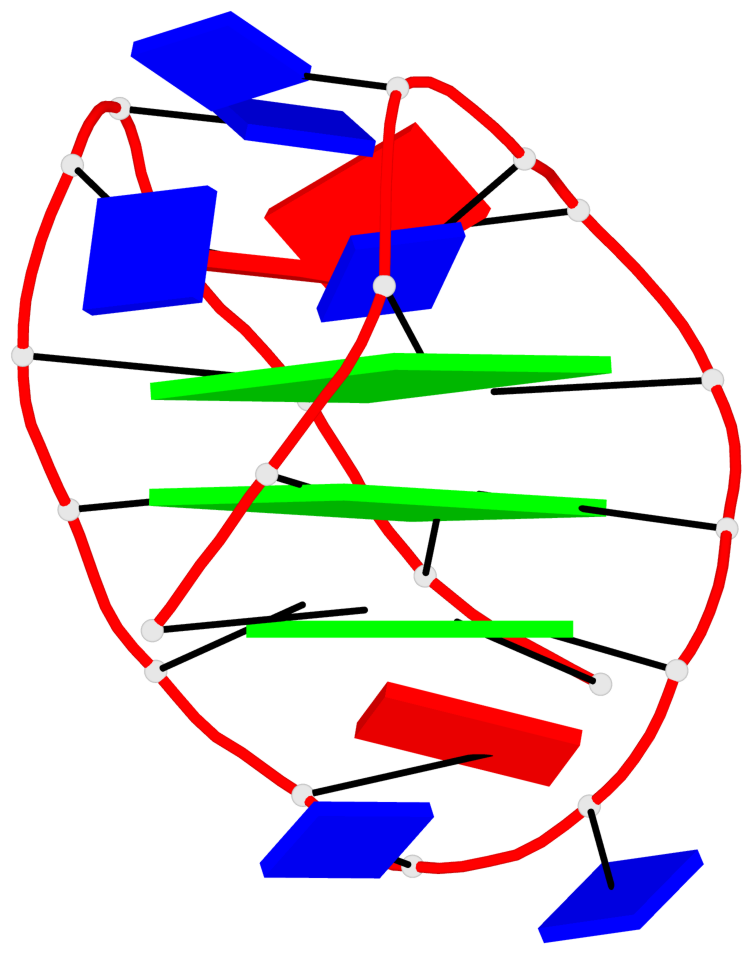
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-LwD+Ln), basket(2+2), UDDU
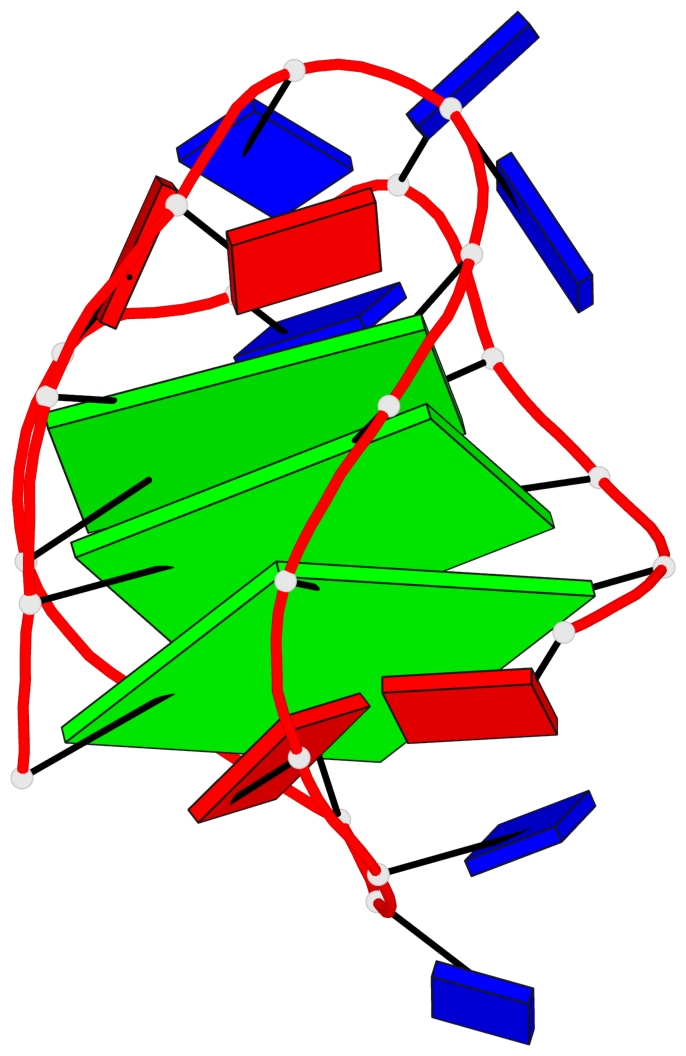
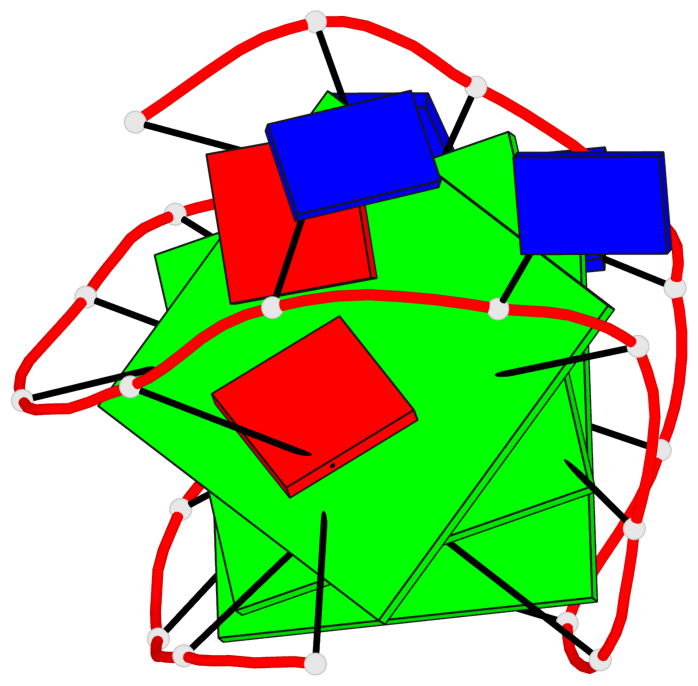
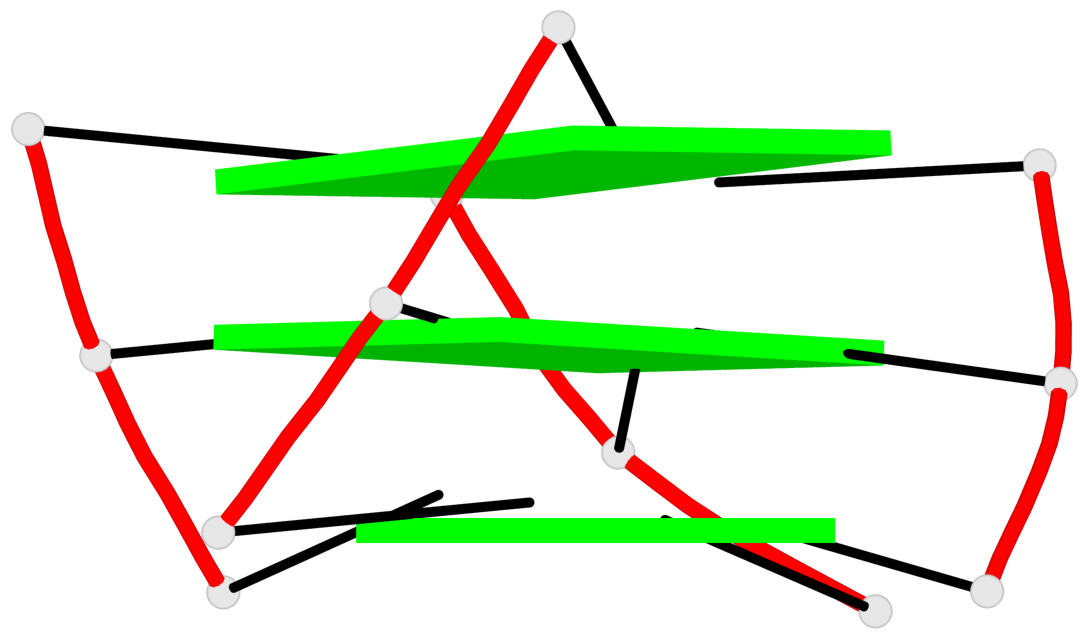
Base-block schematics in six views
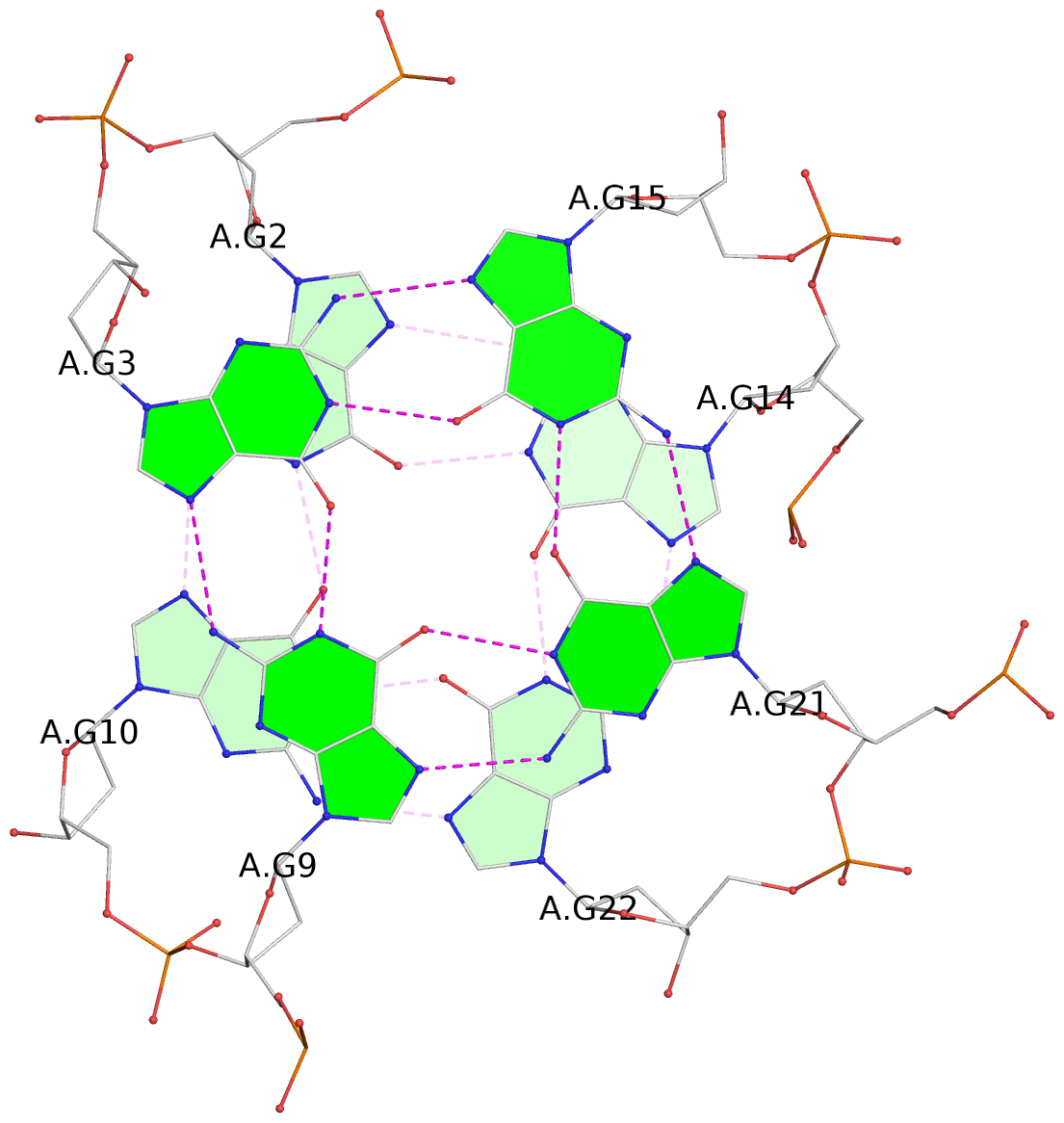
List of 3 G-tetrads
1 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.412 type=other nts=4 GGGG A.DG2,A.DG10,A.DG22,A.DG14 2 glyco-bond=s--s sugar=3--- groove=w-n- planarity=0.275 type=bowl nts=4 GGGG A.DG3,A.DG9,A.DG21,A.DG15 3 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.341 type=other nts=4 GGGG A.DG4,A.DG8,A.DG20,A.DG16
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.