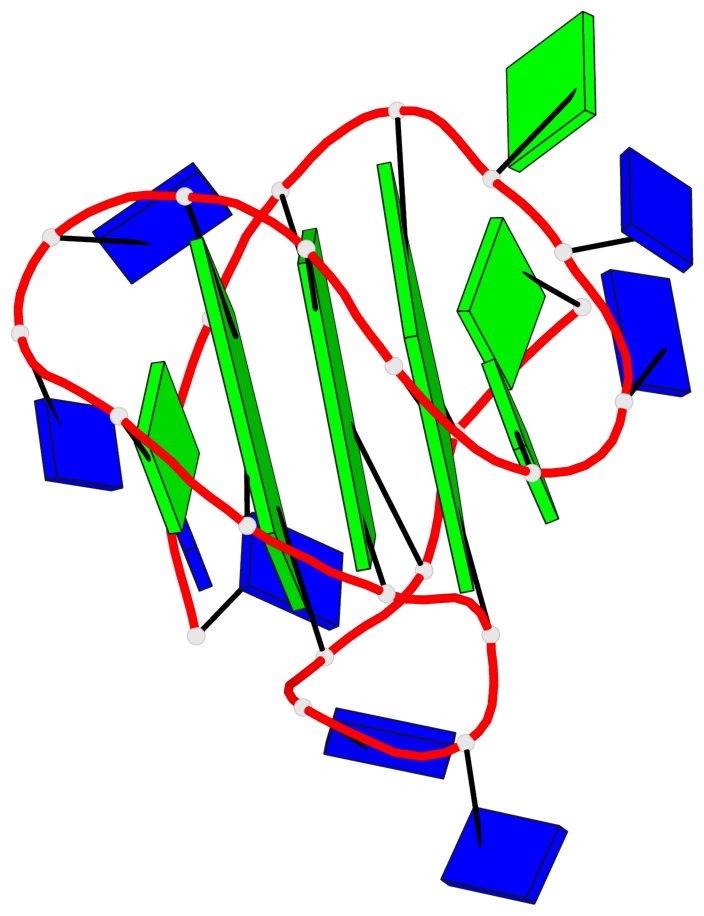
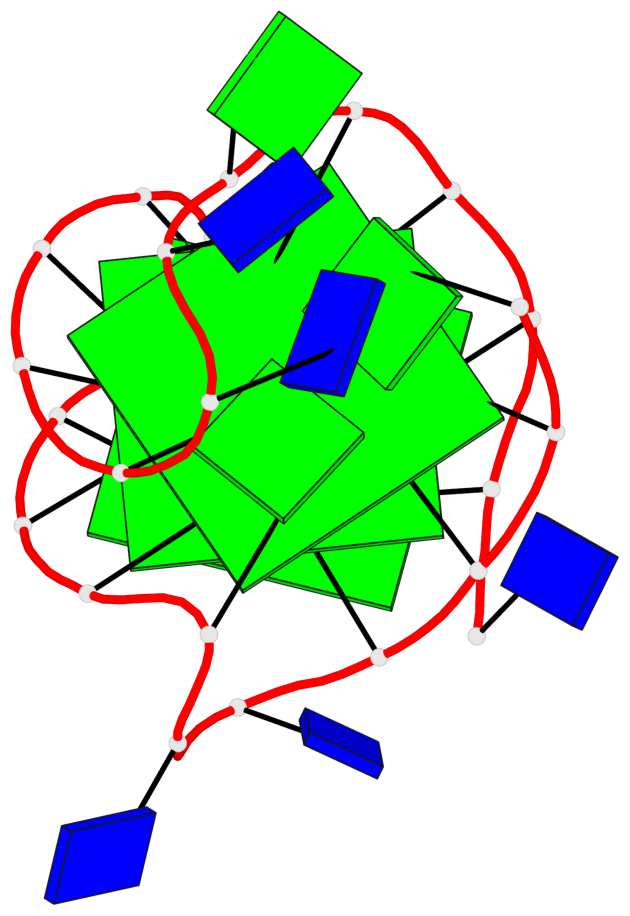
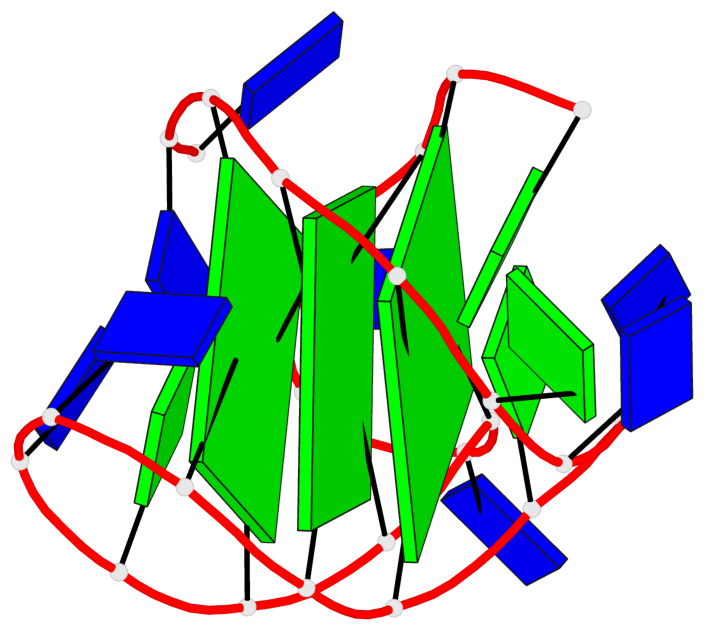
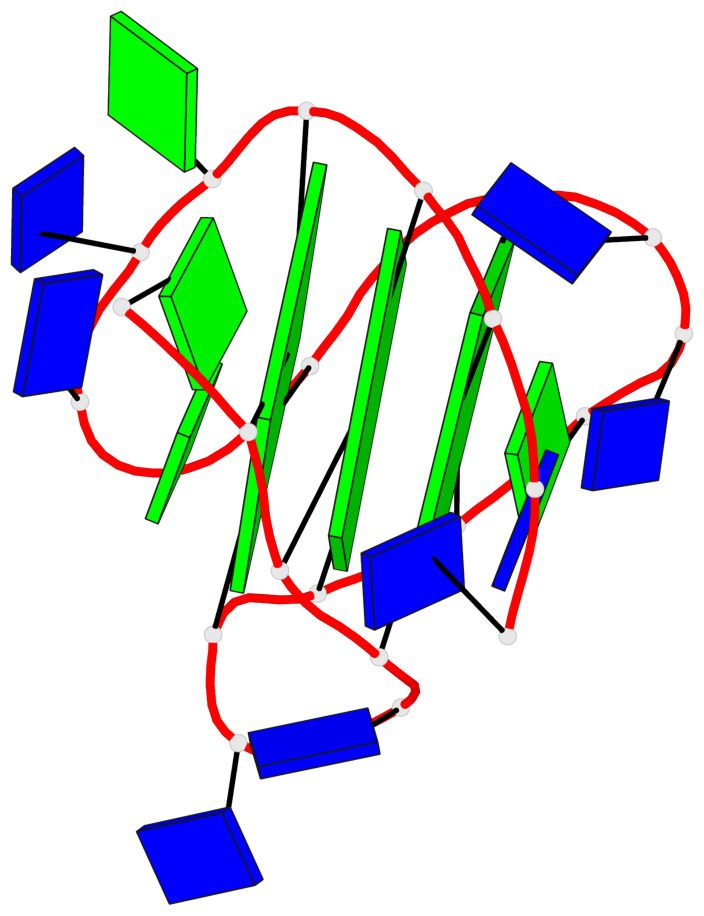
Detailed DSSR results for the G-quadruplex: PDB entry 186d
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 186d
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of the tetrahymena telomeric repeat d(t2g4)4 g-tetraplex
- Reference
- Wang Y, Patel DJ (1994): "Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex." Structure, 2, 1141-1156. doi: 10.1016/S0969-2126(94)00117-0.
- Abstract
- Background: Telomeres in eukaryotic organisms are protein-DNA complexes which are essential for the protection and replication of chromosomal termini. The telomeric DNA of Tetrahymena consists of T2G4 repeats, and models have been previously proposed for the intramolecular folded structure of the d(T2G4)4 sequence based on chemical footprinting and cross-linking data. A high-resolution solution structure of this sequence would allow comparison with the structures of related G-tetraplexes.
Results: The solution structure of the Na(+)-stabilized d(T2G4)4 sequence has been determined using a combined NMR-molecular dynamics approach. The sequence folds intramolecularly into a right-handed G-tetraplex containing three stacked G-tetrads connected by linker segments consisting of a G-T-T-G lateral loop, a central T-T-G lateral loop and a T-T segment that spans the groove through a double chain reversal. The latter T-T connectivity aligns adjacent G-G-G segments in parallel and introduces a new G-tetraplex folding topology with unprecedented combinations of strand directionalities and groove widths, as well as guanine syn/anti distributions along individual strands and around individual G-tetrads.
Conclusions: The four repeat Tetrahymena and human G-tetraplexes, which differ by a single guanine for adenine substitution, exhibit strikingly different folding topologies. The observed structural polymorphism establishes that G-tetraplexes can adopt topologies which project distinctly different groove dimensions, G-tetrad base edges and linker segments for recognition by, and interactions with, other nucleic acids and proteins. - G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
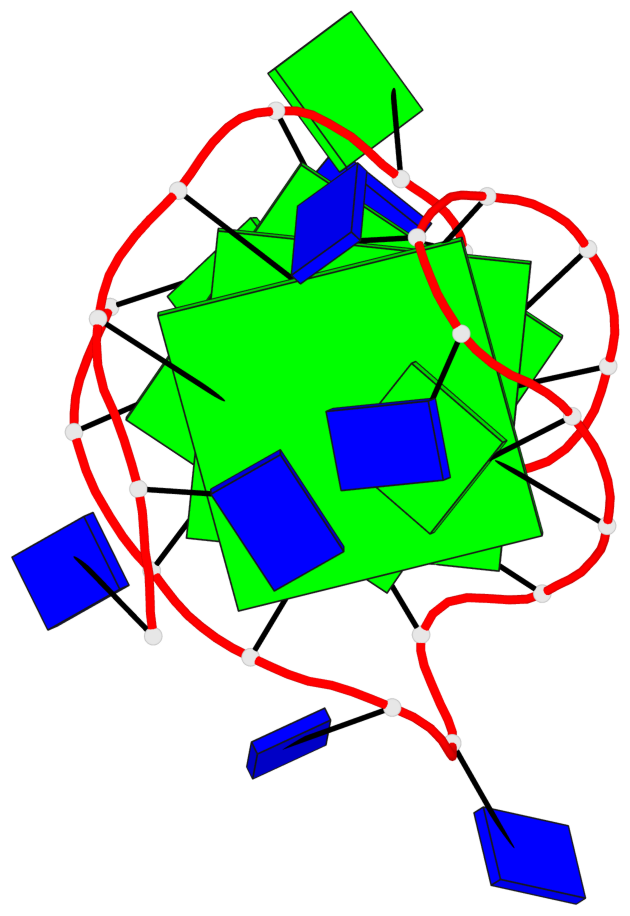
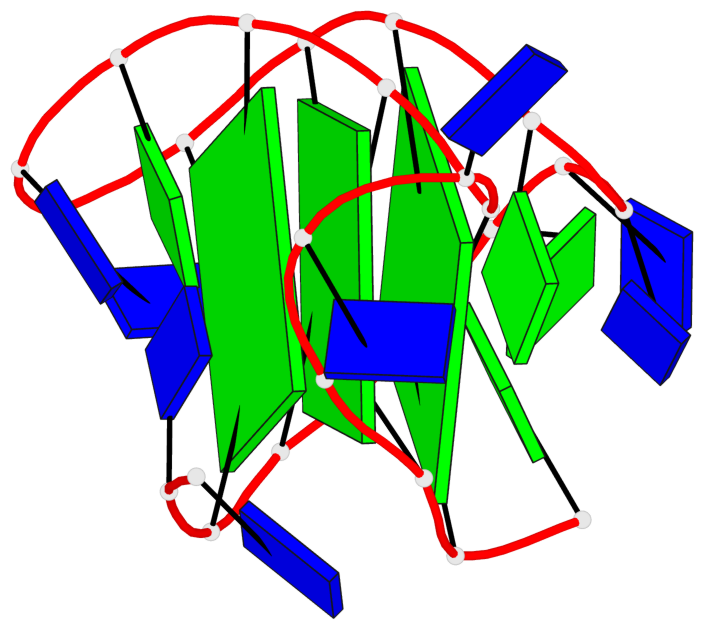
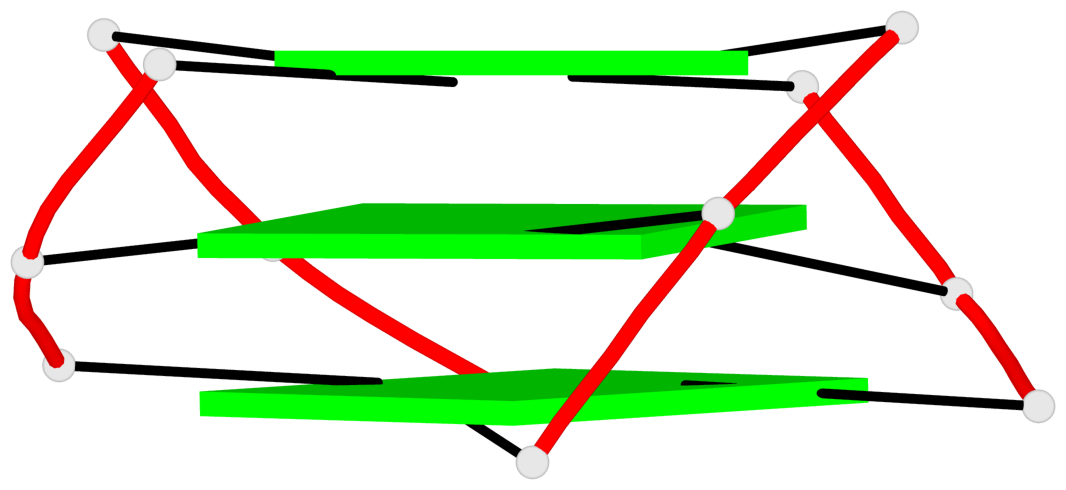
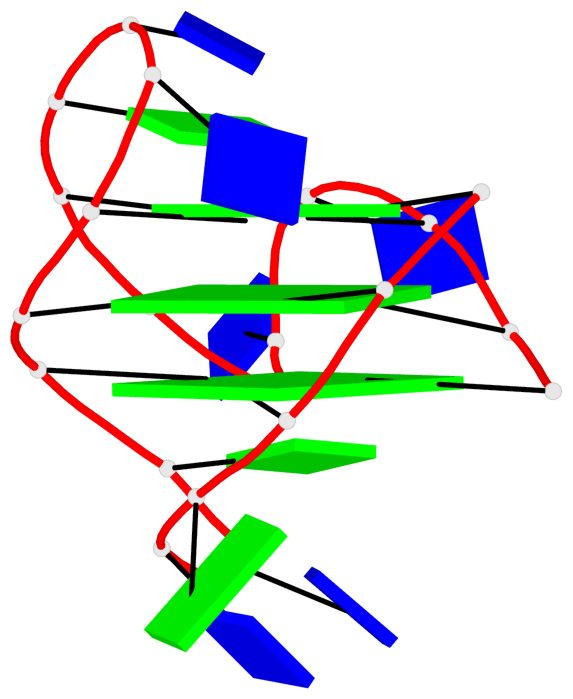
Base-block schematics in six views
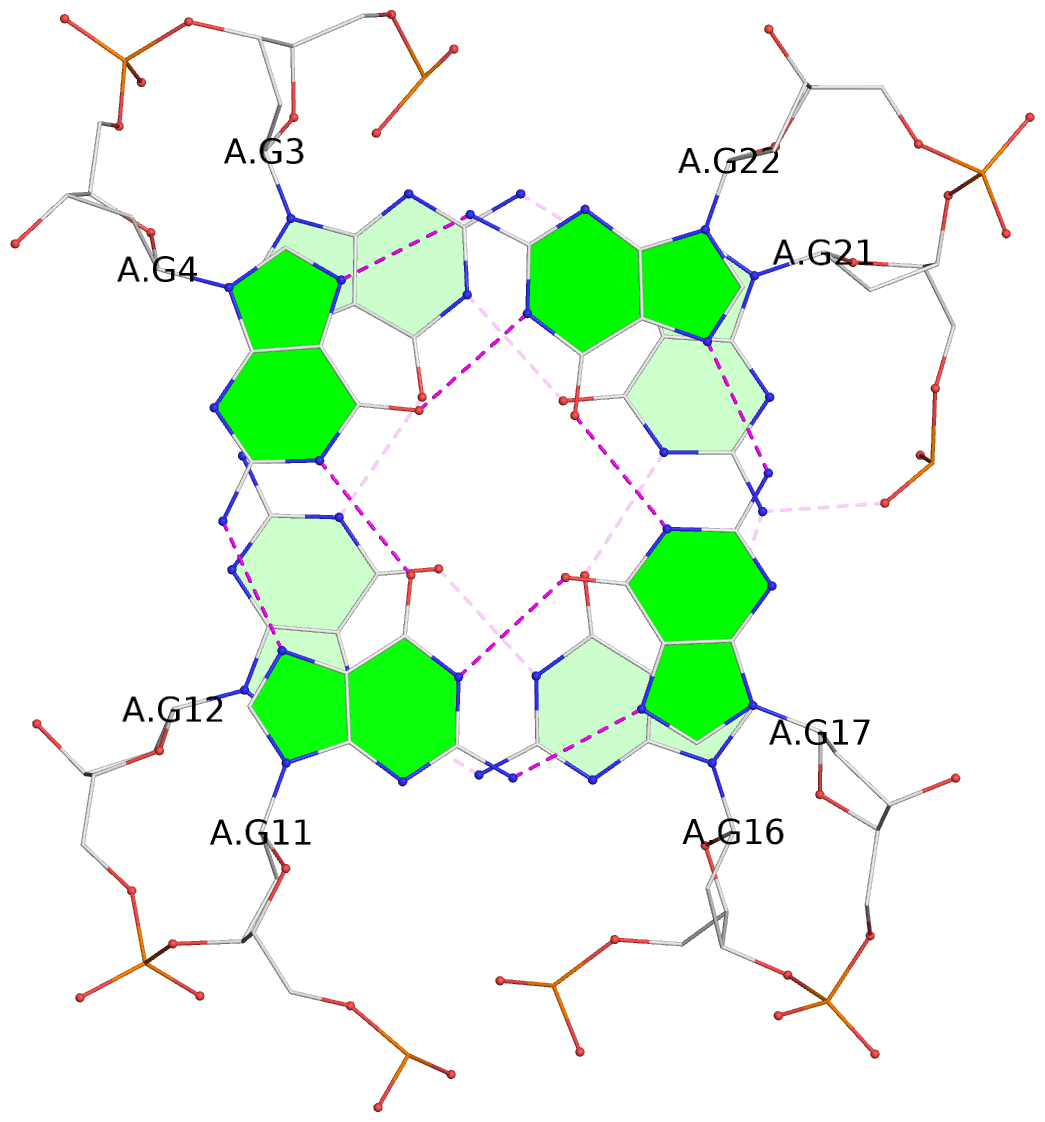
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=---- groove=wn-- planarity=0.418 type=other nts=4 GGGG A.DG3,A.DG12,A.DG16,A.DG21 2 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.445 type=other nts=4 GGGG A.DG4,A.DG11,A.DG17,A.DG22 3 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.333 type=other nts=4 GGGG A.DG5,A.DG10,A.DG18,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.