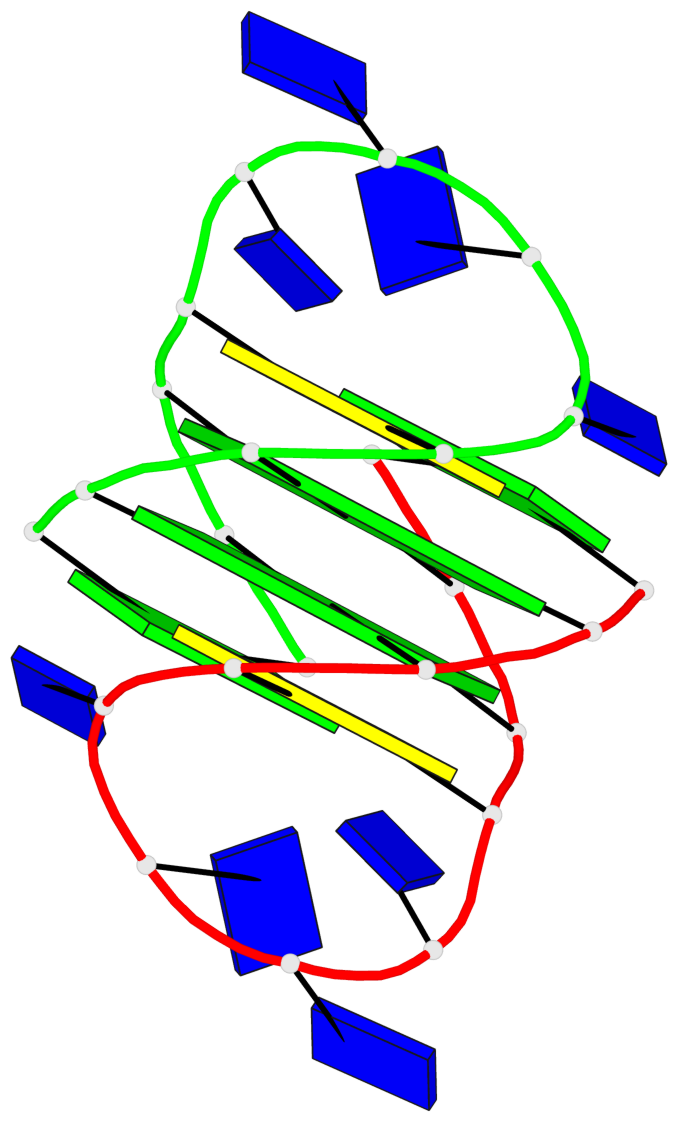
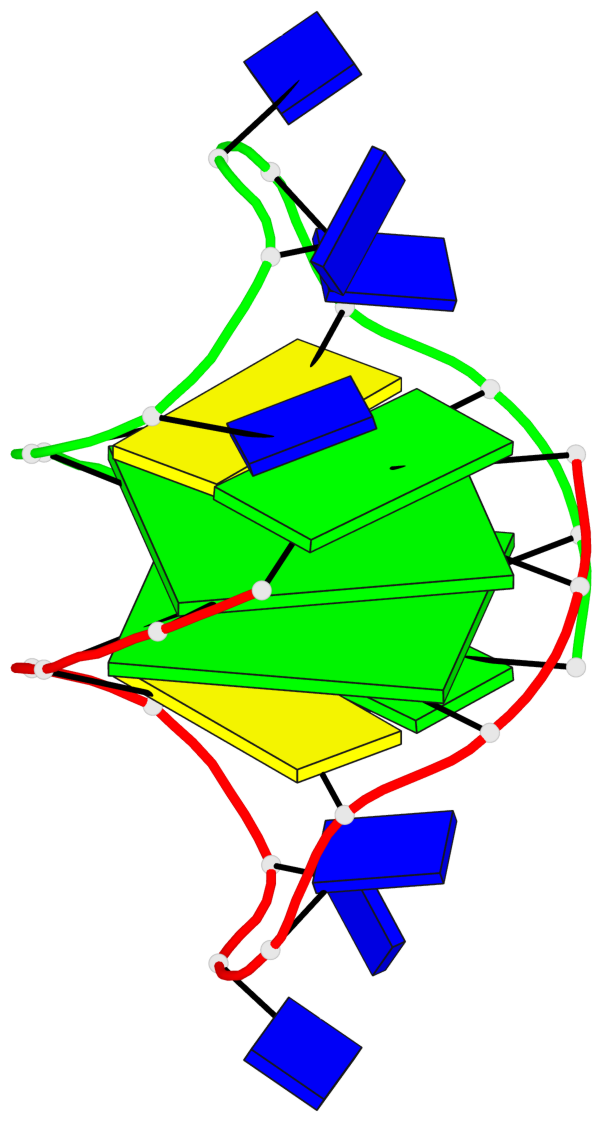
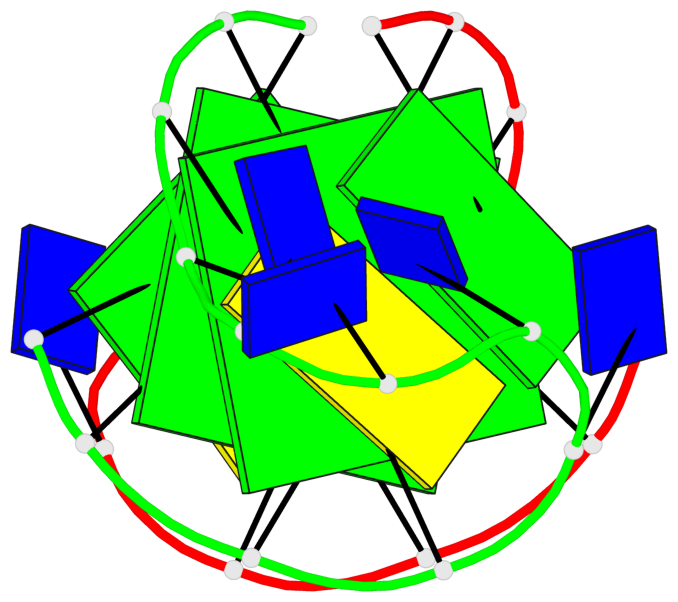
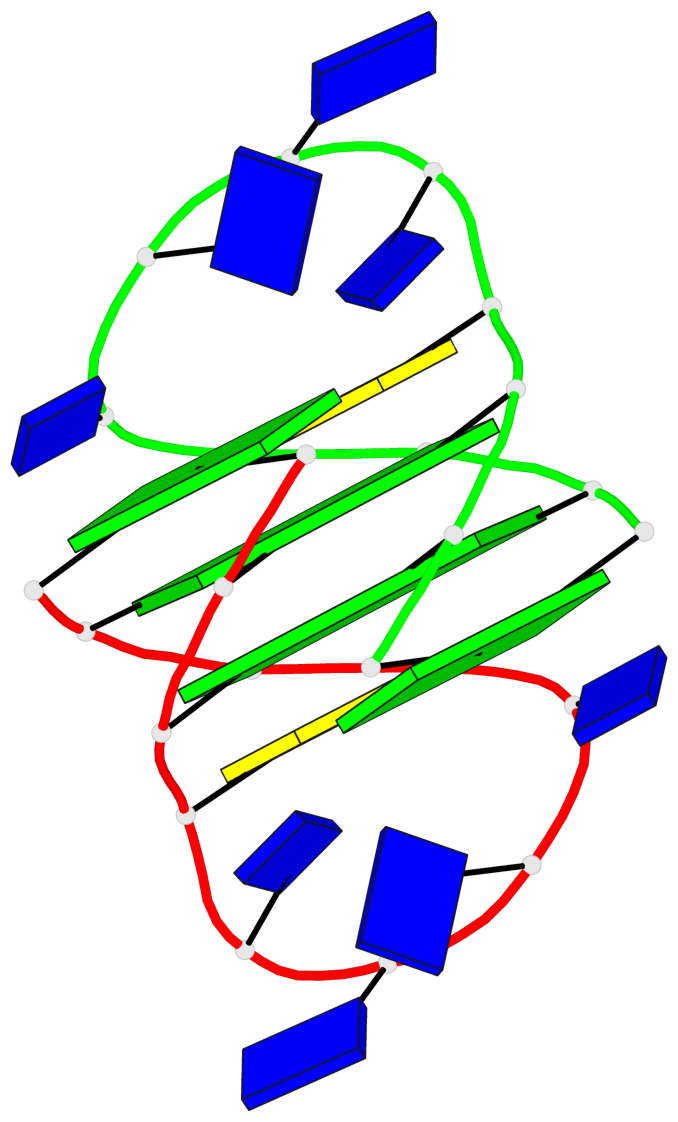
Detailed DSSR results for the G-quadruplex: PDB entry 1a8n
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1a8n
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of a na+ cation stabilized DNA quadruplex containing g.g.g.g and g.c.g.c tetrads formed by g-g-g-c repeats observed in aav and human chromosome 19, NMR, 8 structures
- Reference
- Kettani A, Bouaziz S, Gorin A, Zhao H, Jones RA, Patel DJ (1998): "Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA." J.Mol.Biol., 282, 619-636. doi: 10.1006/jmbi.1998.2030.
- Abstract
- We have applied NMR and molecular dynamics computations including intensity based refinement to define the structure of the d(G-G-G-C-T4-G-G-G-C) dodecanucleotide in 100 mM NaCl solution. The G-G-G-C sequence is of interest since it has been found as tandem repeats in the DNA sequence of human chromosome 19. The same G-G-G-C sequence is also seen as islands in adeno-associated virus, a human parvovirus, which is unique amongst eukaryotic DNA viruses in its ability to integrate site-specifically into a defined region of human chromosome 19. The d(G-G-G-C-T4-G-G-G-C) sequence forms a quadruplex in Na cation containing solution through head-to-tail dimerization of two symmetry-related stem-hairpin loops with adjacent strands antiparallel to each other around the quadruplex. The connecting T4 loops are of the lateral type, resulting in a quadruplex structure containing two internal G.G.G.G tetrads flanked by G.C.G.C tetrads. The G(anti).G(syn).G(anti).G(syn) tetrads are formed through dimerization associated hydrogen bonding alignments of a pair of Hoogsteen G(anti).G(syn) mismatch pairs, while the G(anti).C(anti).G(anti).C(anti) tetrads are formed through dimerization associated bifurcated hydrogen bonding alignments involving the major groove edges of a pair of Watson-Crick G.C base-pairs. The quadruplex contains two distinct narrow and two symmetric wide grooves with extensive stacking between adjacent tetrad planes. The structure of the quadruplex contains internal cavities that can potentially accommodate Na cations positioned between adjacent tetrad planes. Three such Na cations have been modeled into the structure of the d(G-G-G-C-T4-G-G-G-C) quadruplex. Finally, we speculate on the potential role of quadruplex formation involving G.G.G.G and G.C.G.C tetrads during the integration of the adeno-associated parvovirus into its target on human chromosome 19, both of which involve stretches of G-G-G-C sequence elements.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDUD
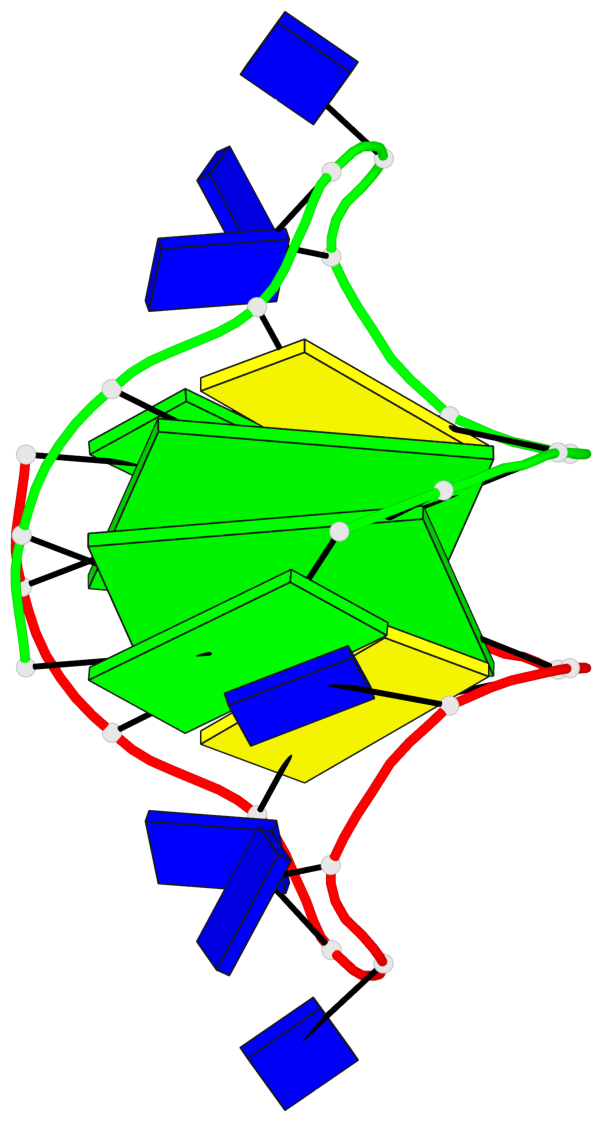
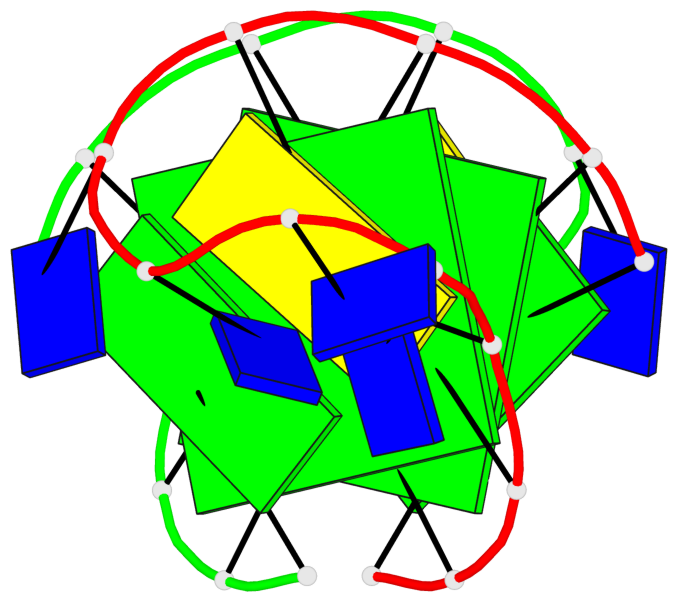
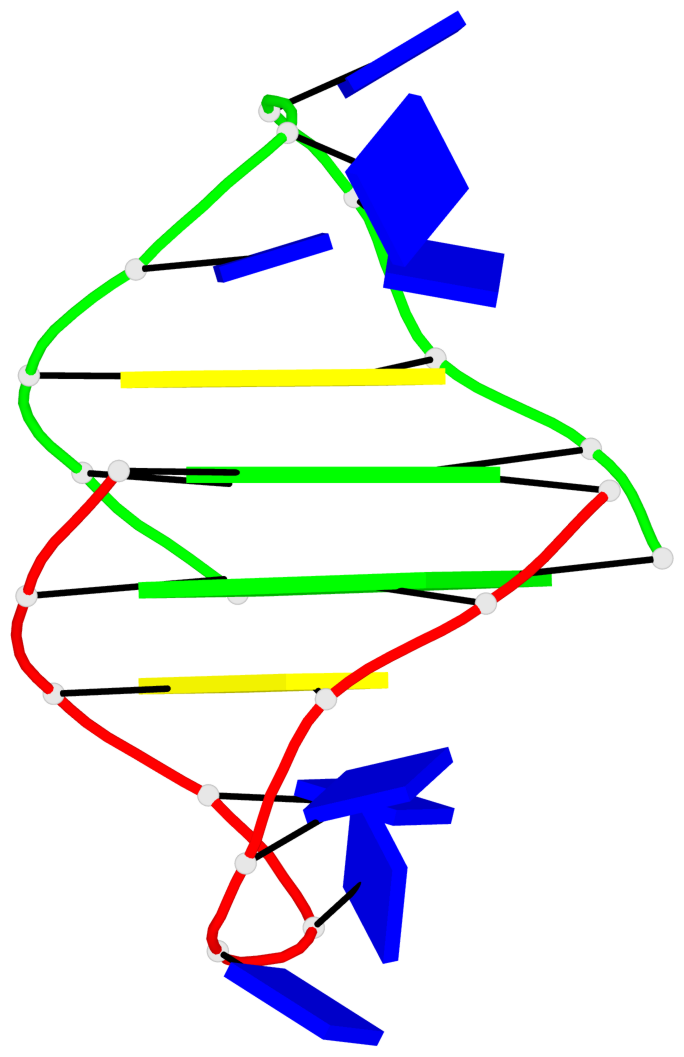
Base-block schematics in six views
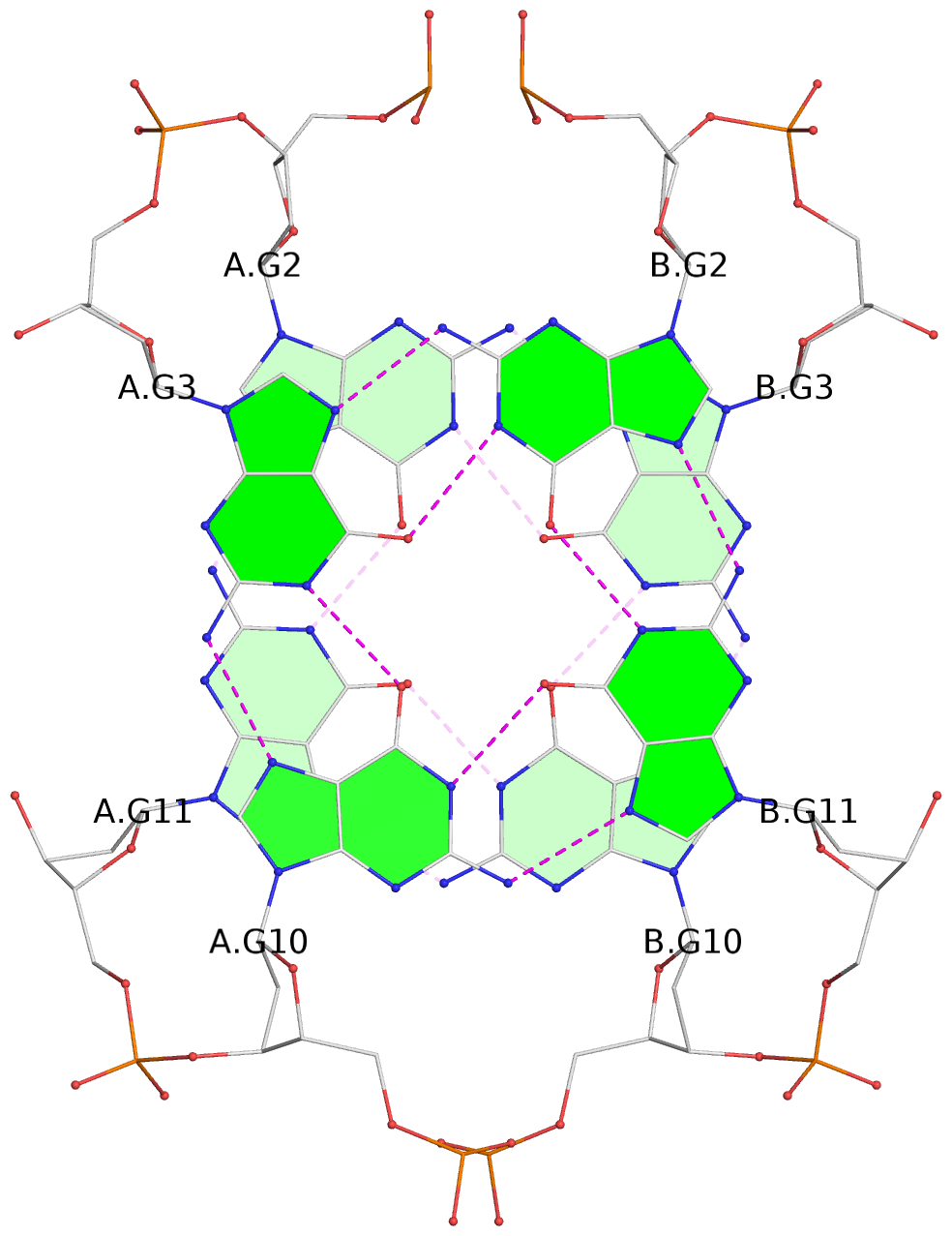
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=.--- groove=wnwn planarity=0.170 type=other nts=4 GGGG A.DG2,A.DG11,B.DG10,B.DG3 2 glyco-bond=-s-s sugar=---. groove=wnwn planarity=0.170 type=other nts=4 GGGG A.DG3,A.DG10,B.DG11,B.DG2
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.