Detailed DSSR results for the G-quadruplex: PDB entry 1bub
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1bub
- Class
- DNA
- Method
- NMR
- Summary
- Determination of internuclear angles of DNA using paramagnetic assisted magnetic alignment
- Reference
- Beger RD, Marathias VM, Volkman BF, Bolton PH (1998): "Determination of internuclear angles of DNA using paramagnetic-assisted magnetic alignment." J.Magn.Reson., 135, 256-259. doi: 10.1006/jmre.1998.1527.
- Abstract
- Paramagnetic ions have been used to assist the magnetic alignment of DNA. The anisotropy of the binding sites is sufficient to give rise to significant alignment of the DNA with the observed proton-carbon dipolar couplings spanning a 70-Hz range. The dipolar couplings have been used to determine the positions of the axial and rhombic alignment axes. The positions of the alignment axes relative to the positions of the binding sites of the paramagnetic europium ions have also been determined.
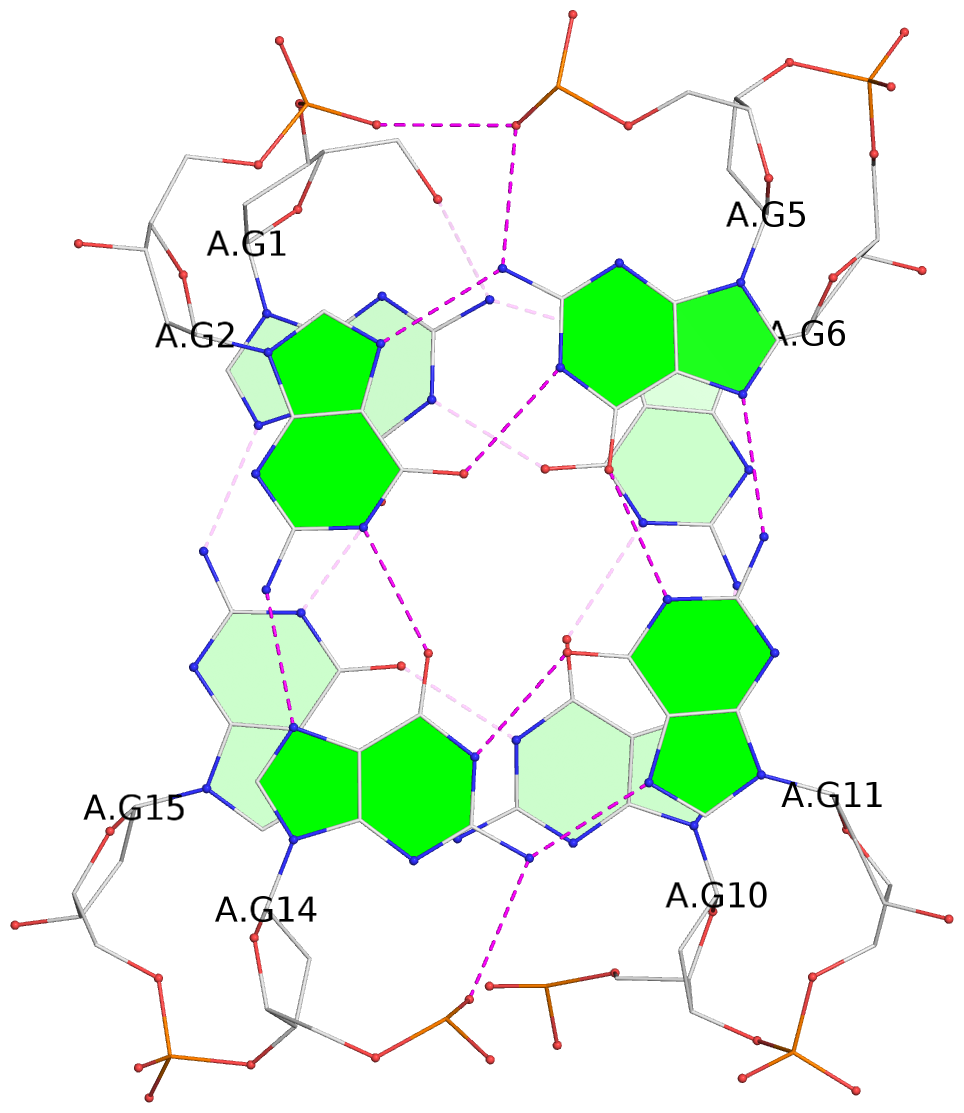
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+Ln+Lw+Ln), chair(2+2), UDUD
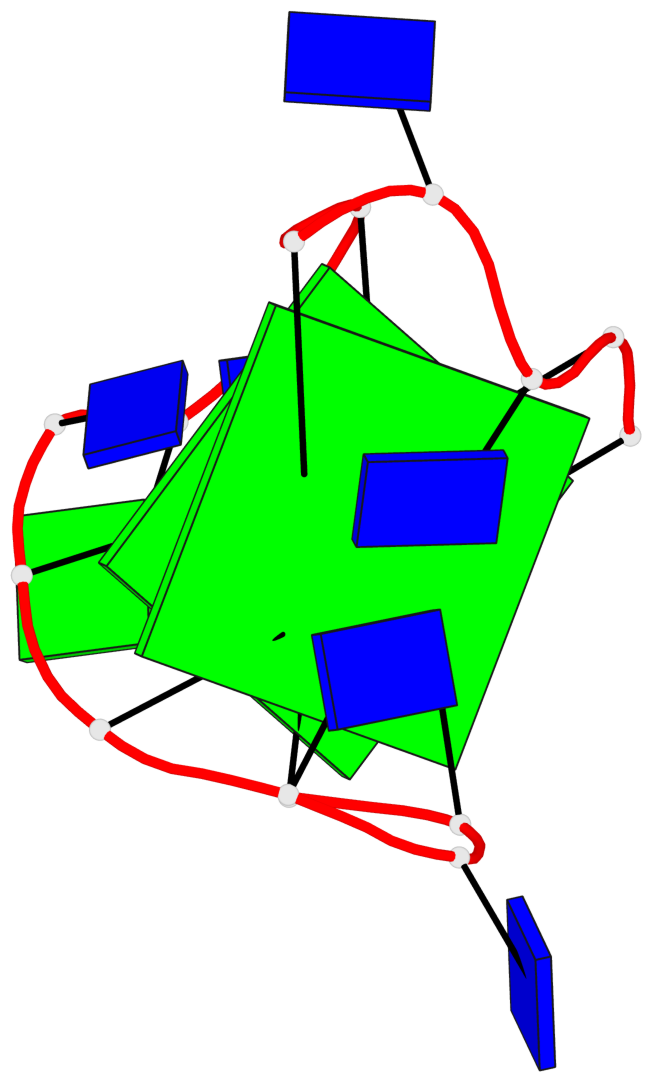
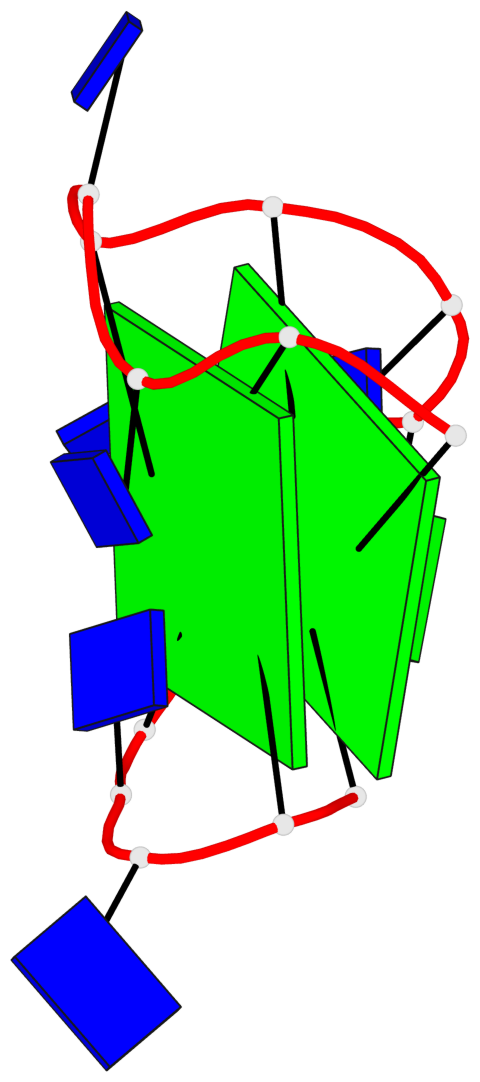
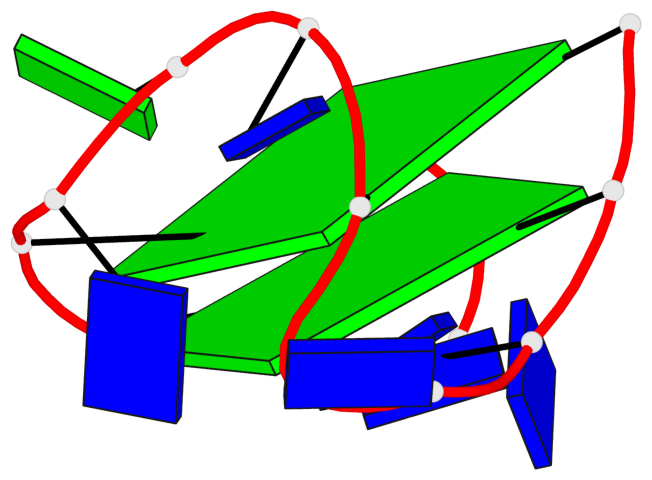
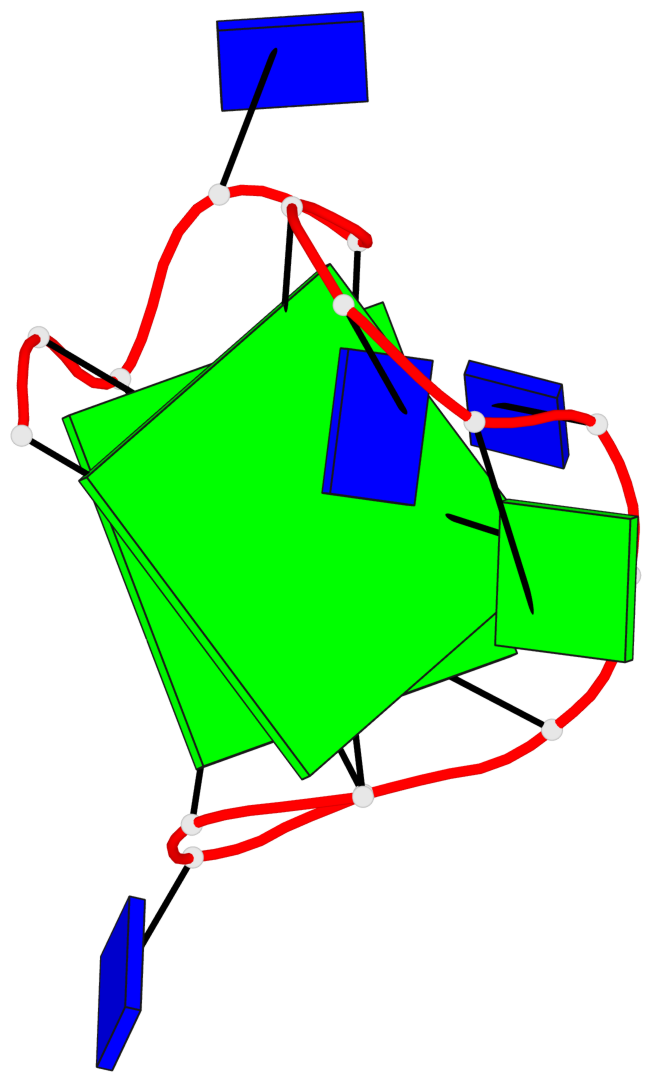
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=3.-. groove=wnwn planarity=0.443 type=bowl nts=4 GGGG A.DG1,A.DG15,A.DG10,A.DG6 2 glyco-bond=-s-s sugar=.-.- groove=wnwn planarity=0.179 type=other nts=4 GGGG A.DG2,A.DG14,A.DG11,A.DG5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.