Detailed DSSR results for the G-quadruplex: PDB entry 1d59
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1d59
- Class
- DNA
- Method
- X-ray (2.3 Å)
- Summary
- Crystal structure of 4-stranded oxytricha telomeric DNA
- Reference
- Kang C, Zhang X, Ratliff R, Moyzis R, Rich A (1992): "Crystal structure of four-stranded Oxytricha telomeric DNA." Nature, 356, 126-131. doi: 10.1038/356126a0.
- Abstract
- The sequence d(GGGGTTTTGGGG) from the 3' overhang of the Oxytricha telomere has been crystallized and its three-dimensional structure solved to 2.5 A resolution. The oligonucleotide forms hairpins, two of which join to make a four-stranded helical structure with the loops containing four thymine residues at either end. The guanine residues are held together by cyclic hydrogen bonding and an ion is located in the centre. The four guanine residues in each segment have a glycosyl conformation that alternates between anti and syn. There are two four-stranded molecules in the asymmetric unit showing that the structure has some intrinsic flexibility.
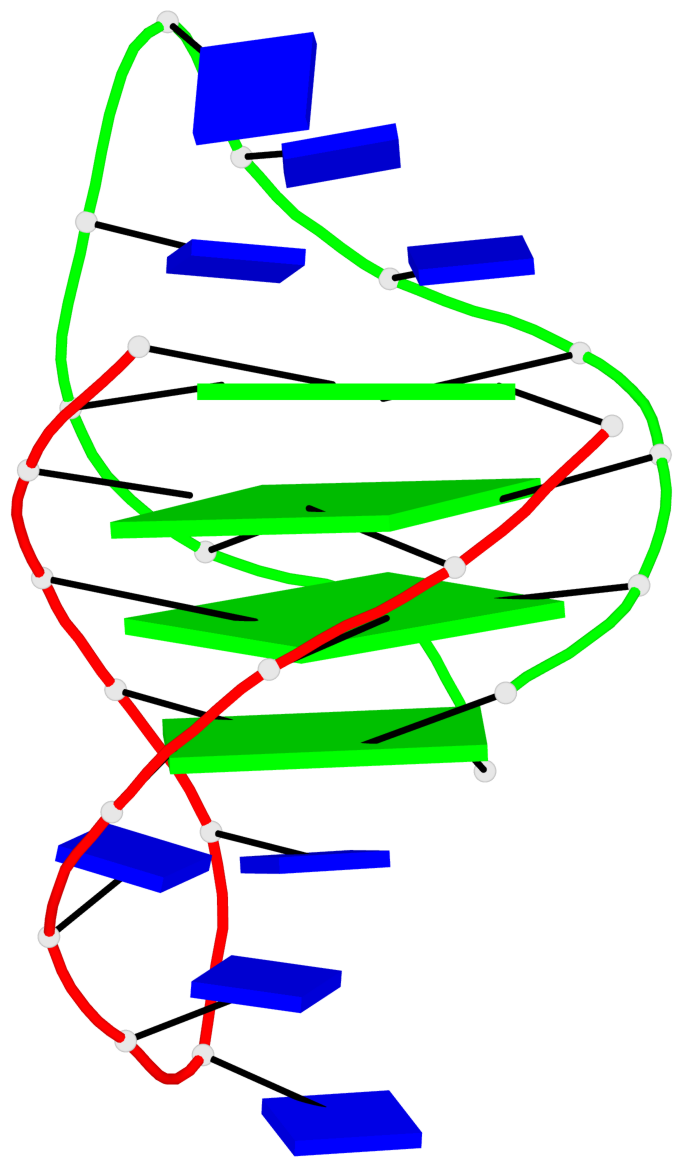
- G4 notes
- 8 G-tetrads, 2 G4 helices, 2 G4 stems, (2+2), UDUD
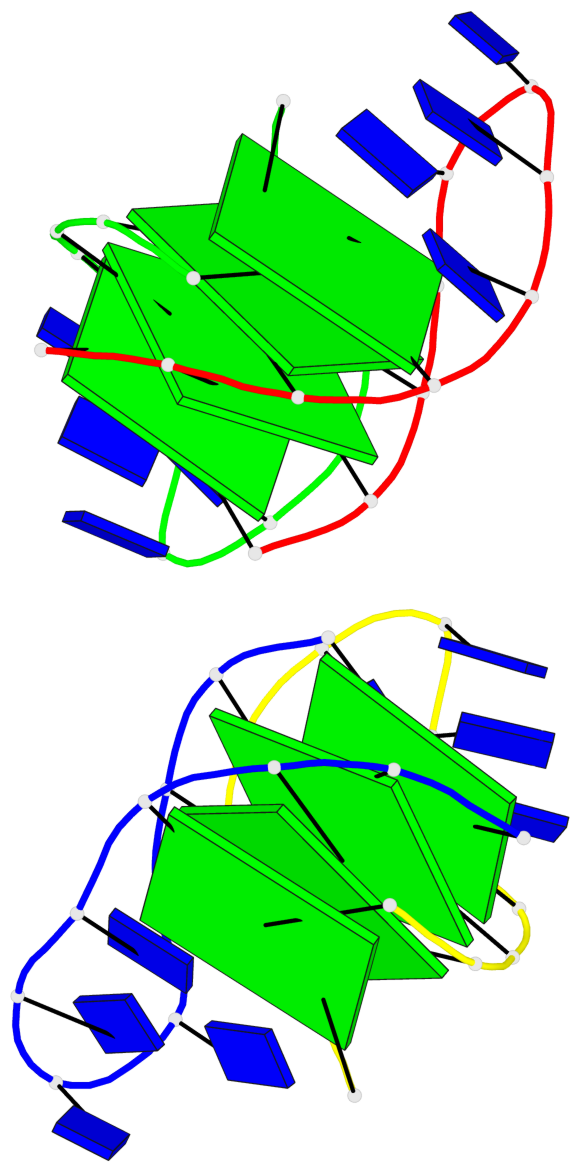
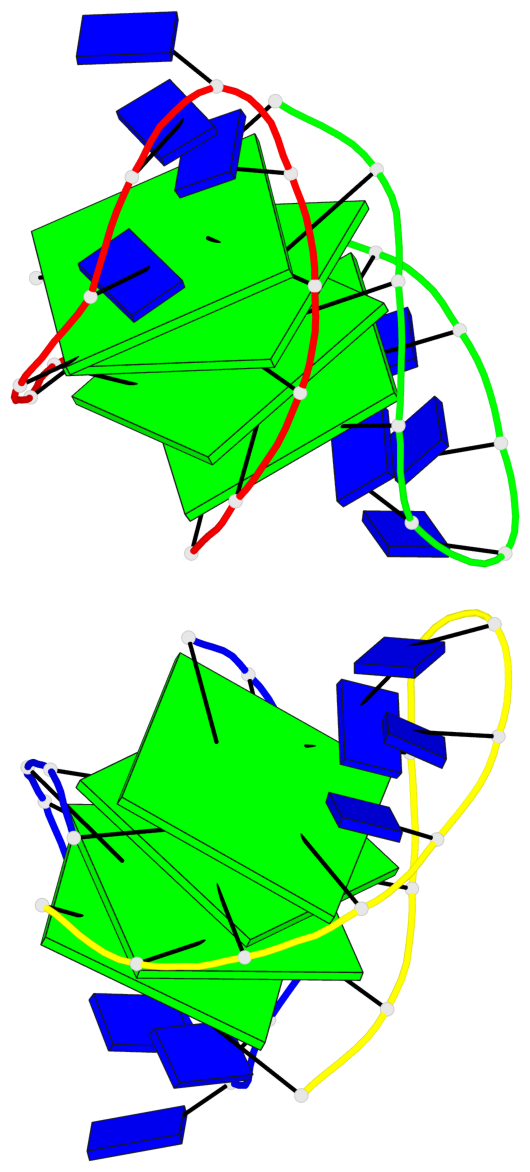
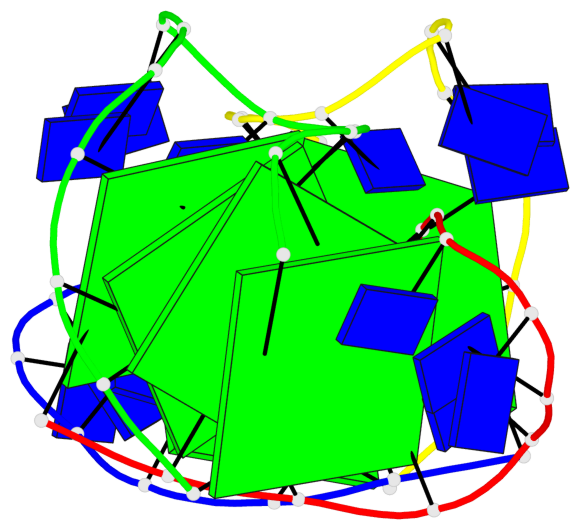
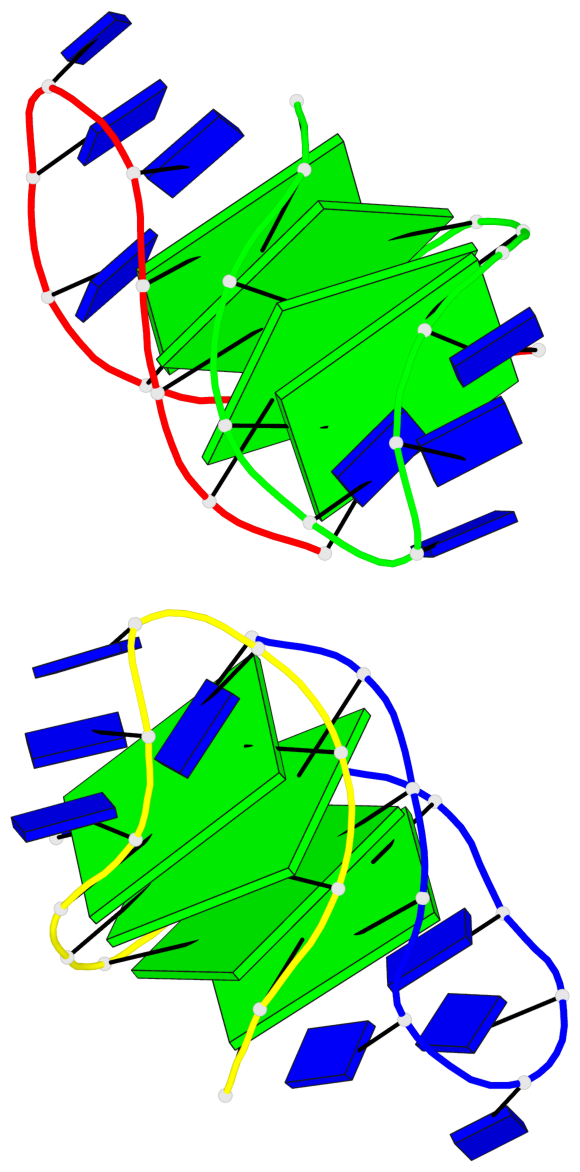
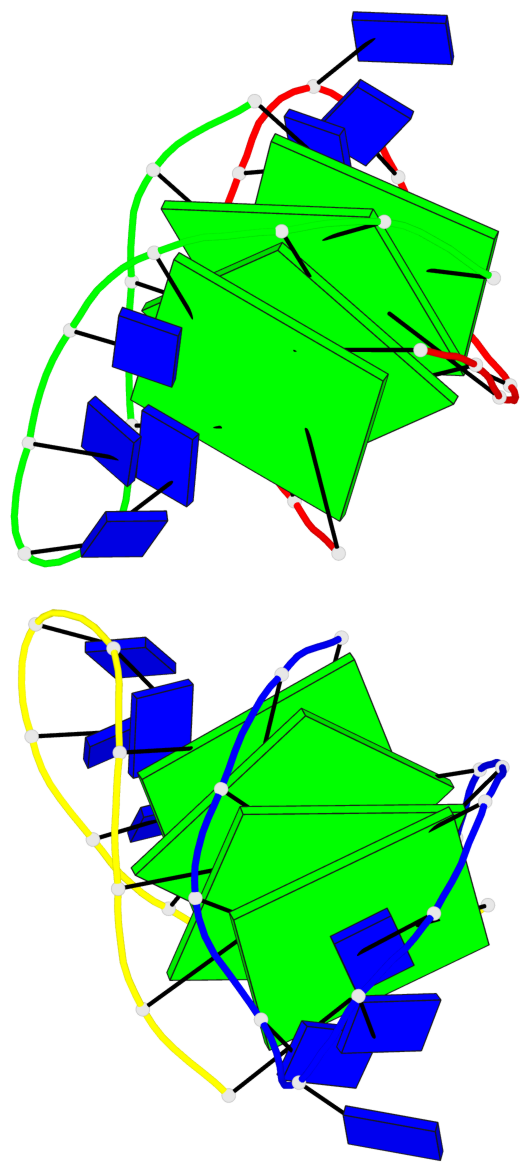
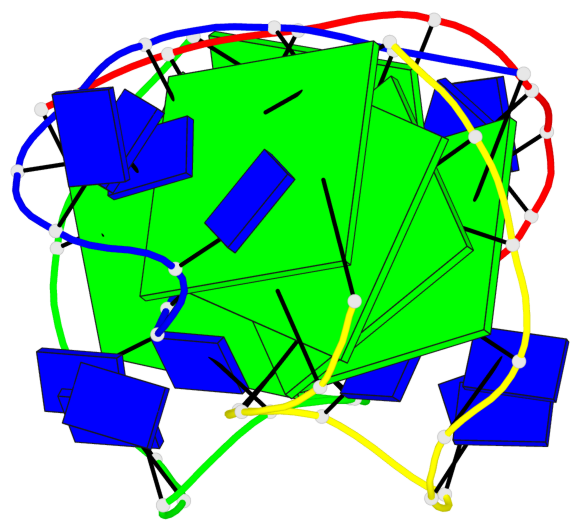
Base-block schematics in six views
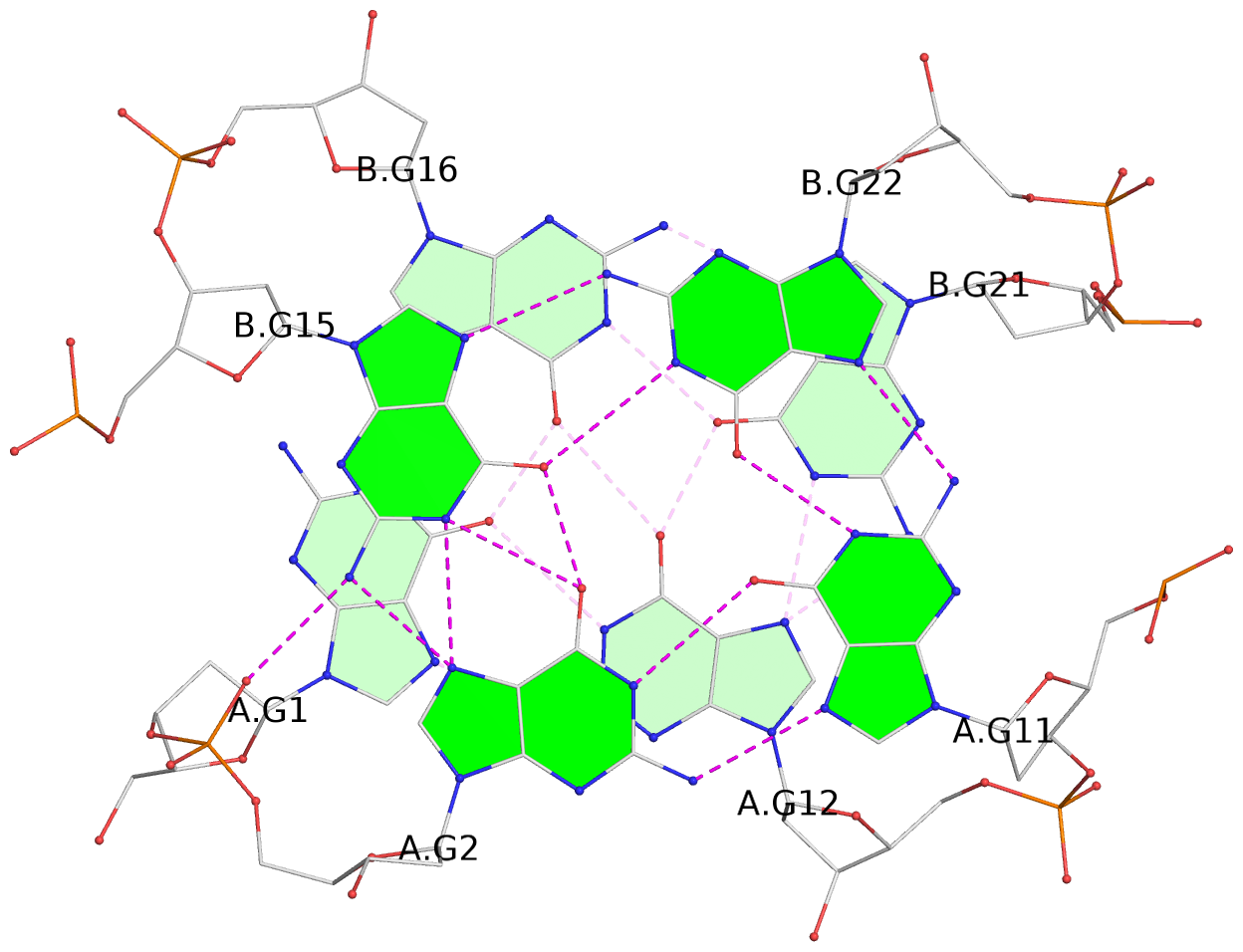
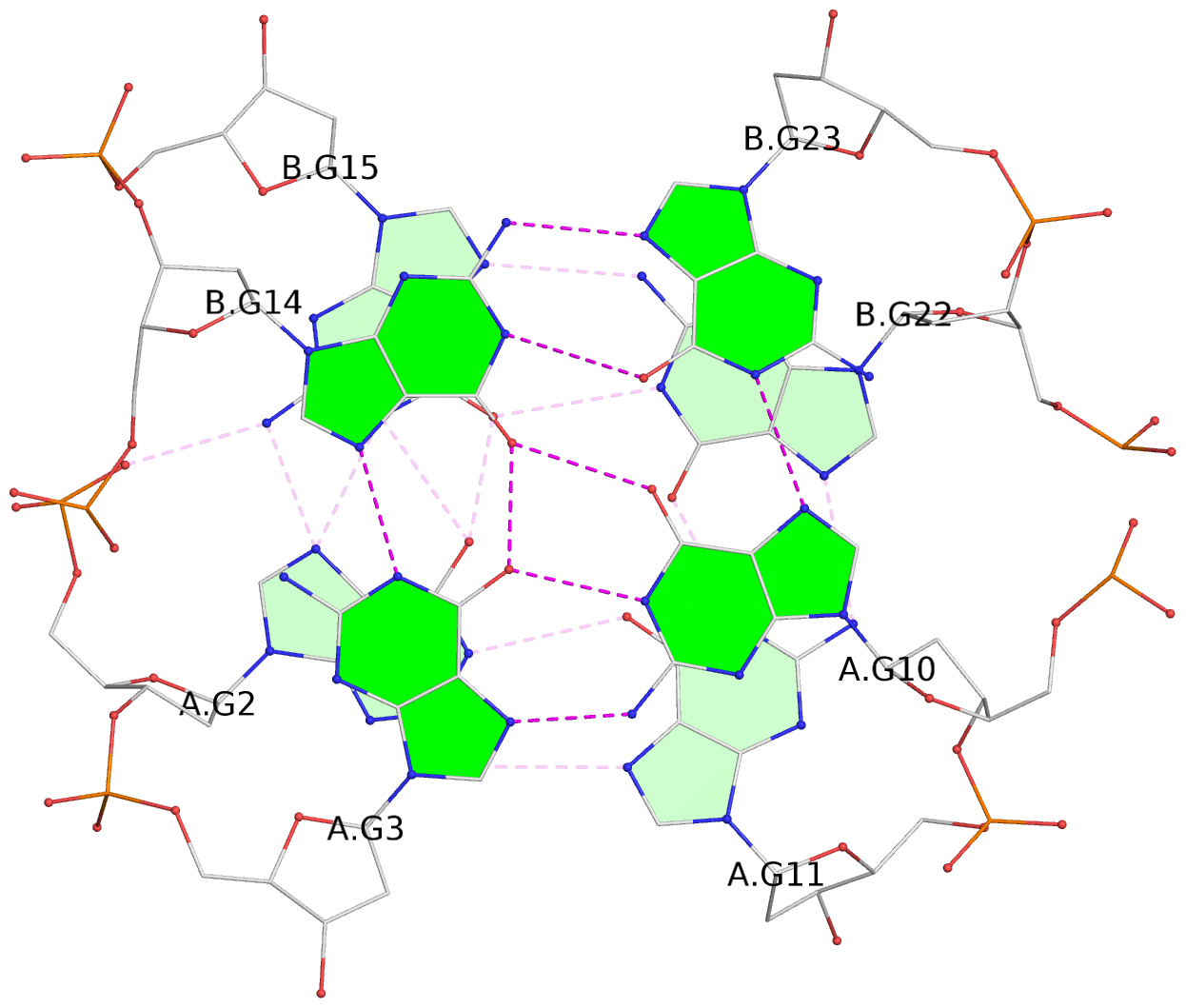
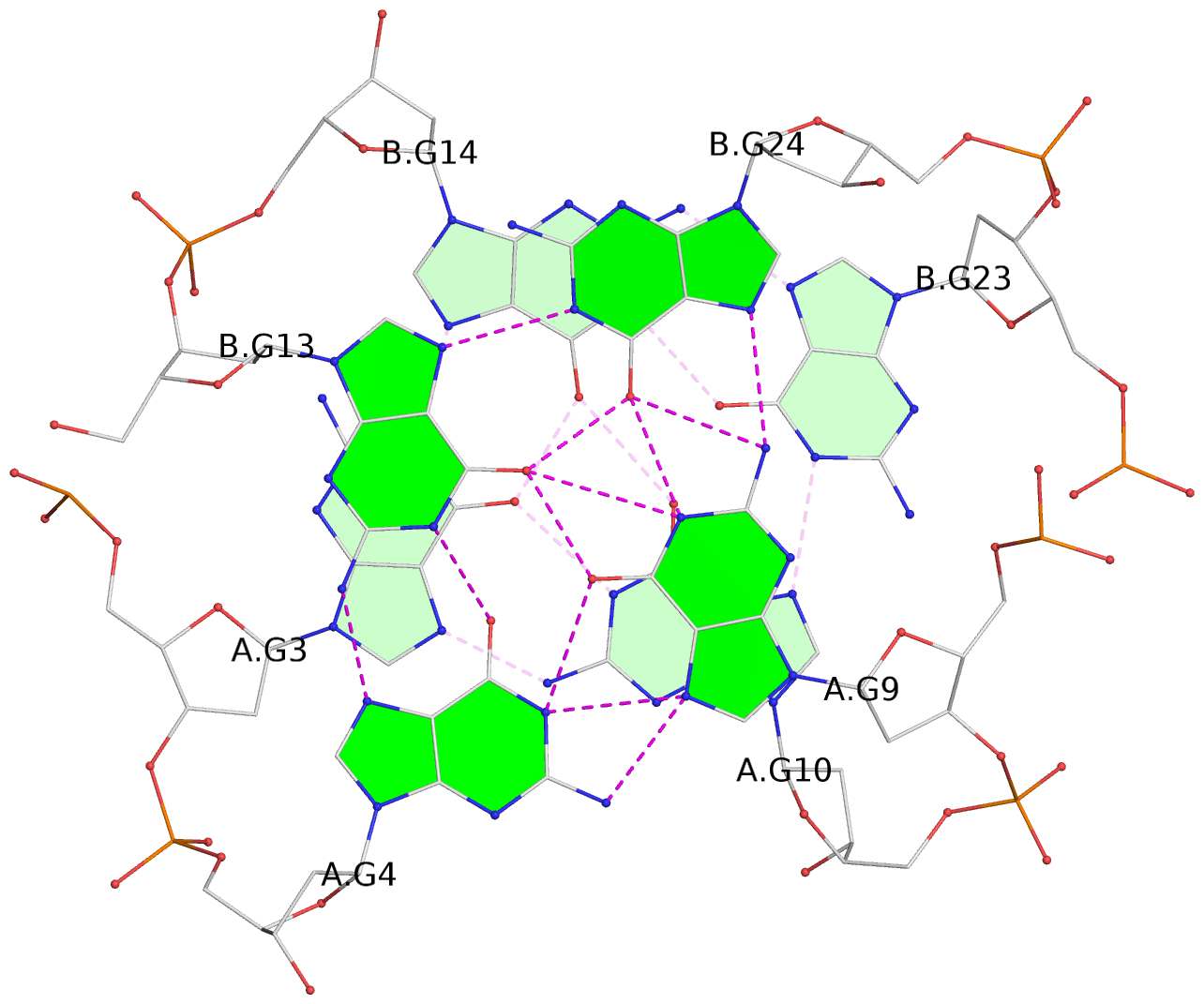
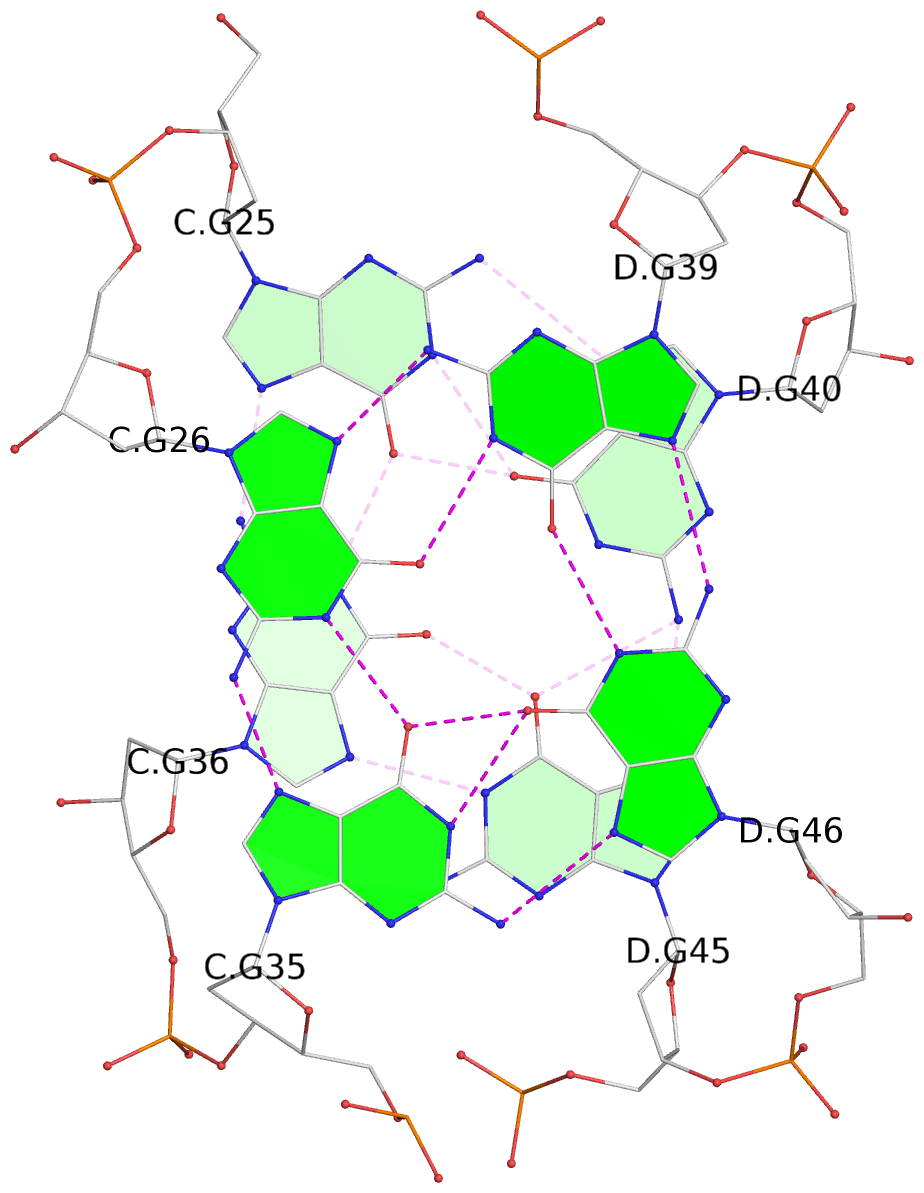
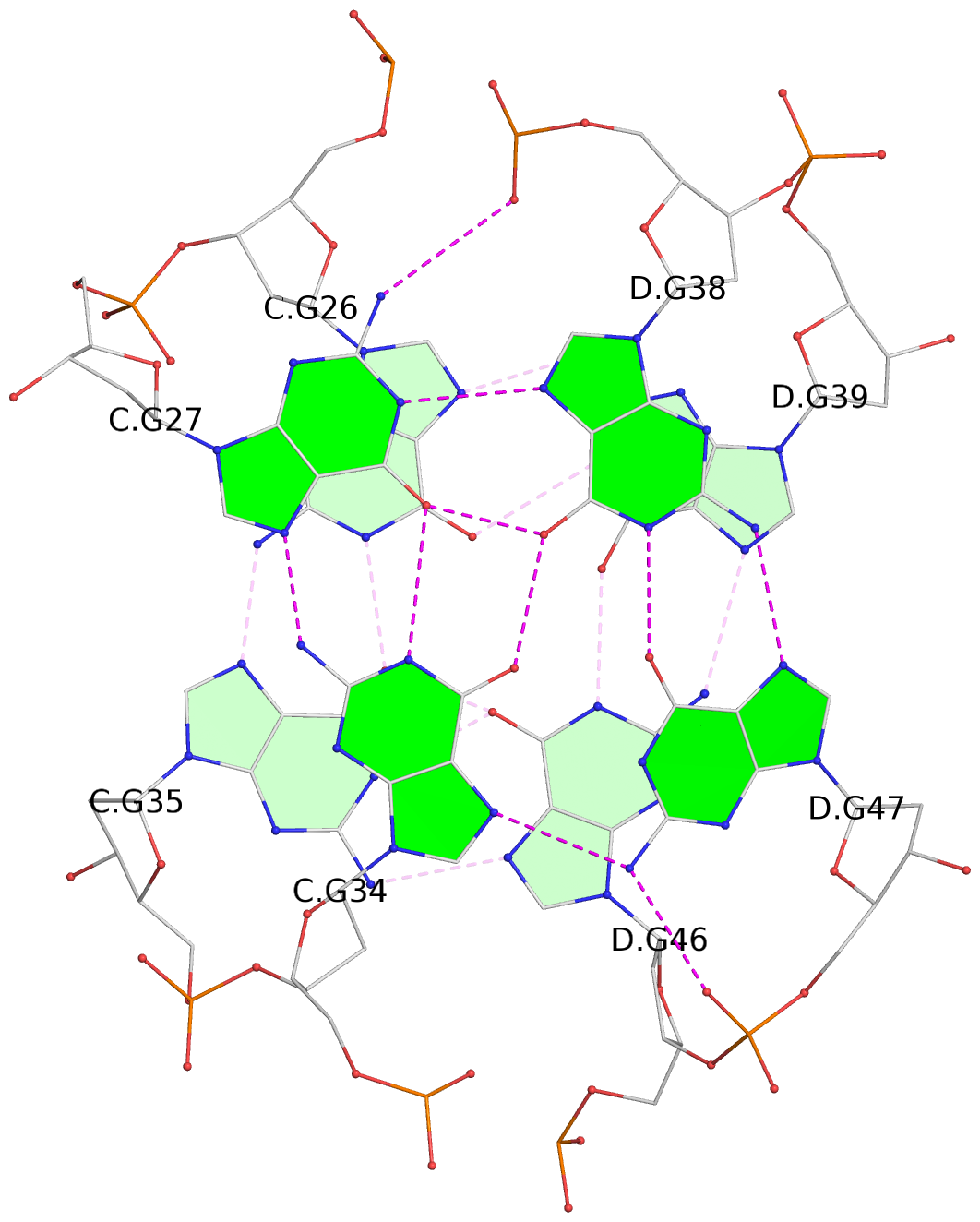
List of 8 G-tetrads
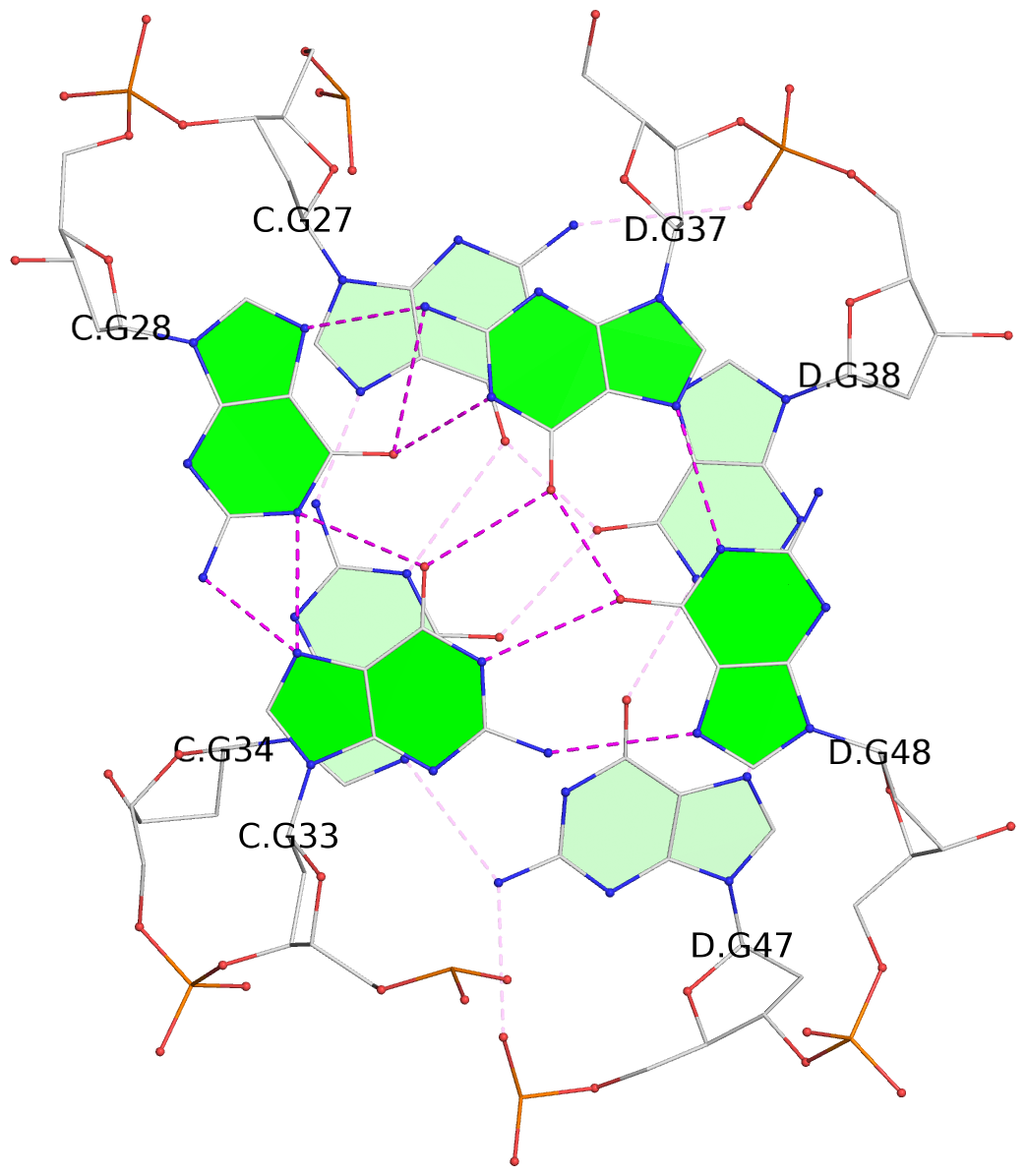
1 glyco-bond=--s- sugar=-.-- groove=-wn- planarity=0.244 type=other nts=4 GGGG A.DG1,B.DG16,B.DG21,A.DG12 2 glyco-bond=-s-s sugar=33.. groove=wnwn planarity=0.284 type=other nts=4 GGGG A.DG2,A.DG11,B.DG22,B.DG15 3 glyco-bond=--s- sugar=.-3- groove=-wn- planarity=0.425 type=other nts=4 GGGG A.DG3,B.DG14,B.DG23,A.DG10 4 glyco-bond=-s-s sugar=-.3- groove=wnwn planarity=0.374 type=other nts=4 GGGG A.DG4,A.DG9,B.DG24,B.DG13 5 glyco-bond=s-s- sugar=-3-3 groove=wnwn planarity=0.320 type=other nts=4 GGGG C.DG25,C.DG36,D.DG45,D.DG40 6 glyco-bond=-s-s sugar=-3.3 groove=wnwn planarity=0.323 type=other nts=4 GGGG C.DG26,C.DG35,D.DG46,D.DG39 7 glyco-bond=s-s- sugar=--33 groove=wnwn planarity=0.441 type=bowl nts=4 GGGG C.DG27,C.DG34,D.DG47,D.DG38 8 glyco-bond=-s-s sugar=.--- groove=wnwn planarity=0.373 type=other nts=4 GGGG C.DG28,C.DG33,D.DG48,D.DG37
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.